Identification of a promoter region generating Sry circular transcripts both in germ cells from male...
-
Upload
independent -
Category
Documents
-
view
5 -
download
0
Transcript of Identification of a promoter region generating Sry circular transcripts both in germ cells from male...
BIOLOGY OF REPRODUCTION 57, 1128-1135 (1997)
Identification of a Promoter Region Generating Sry Circular Transcripts Both inGerm Cells from Male Adult Mice and in Male Mouse Embryonal Gonads'
Susanna Dolci, Paola Grimaldi, Raffaele Geremia, Maurizio Pesce, and Pellegrino Rossi2
Dipartimento di Sanita' Pubblica e Biologia Cellulare, Universita' di Roma "Tor Vergata," Rome, Italy
ABSTRACT
The mouse testis determining gene Sry is expressed in so-matic cells of the differentiating male gonad as a linear tran-script, encoding a transcription factor containing an HMG box.In the adult mouse testis, Sry expression occurs in meiotic andpostmeiotic germ cells. The mouse genomic Sry locus is char-acterized by two arms of a large inverted repeat, flanking aunique region that, between an acceptor and a donor splice site,contains a single exon encoding the Sry protein. In germ cellsfrom the adult mouse testis, Sry RNA is a circular molecule,which is generated by an inverted splicing event that utilizes theabove-mentioned splice sites. Thus, a circular exon is spliced outstarting from a large linear RNA precursor containing both armsof the inverted repeat, which pair and generate a large stem-loop structure.
Using reverse transcription-polymerase chain reaction and anRNase protection assay, we have now mapped the 5' end of thisprecursor RNA in the 5' arm of the inverted repeat. Gel mobilityshift assay and in vitro transcription with nuclear extracts fromadult germ cells further confirm that a region immediately 5'upstream of two transcriptional initiation sites of the precursorRNA contains a promoter sequence in which two consensus Srybinding sequences are specifically recognized by nuclear factorspresent in adult germ cells but not in Sertoli cells. We also showthat the linear precursor of the Sry circular transcript and itssplicing product are specifically expressed not only in adultgerm cells but also in male embryonal gonads between 11.5 and13.5 days postcoitum, immediately after the expression of thelinear transcript starting from the unique region.
INTRODUCTION
Testis determination in mammals is a cascade of eventsthat are controlled by the master regulatory gene Sry lo-cated on the Y chromosome [1, 2]. Sry expression in thesomatic cell compartment of the undifferentiated male em-bryonic gonad leads to testicular cord formation and sub-sequent male phenotype [3]. This is achieved through dif-ferentiation of Sertoli and Leydig cells instead of follicularcells, with subsequent proper hormonal production. Sry ex-pression appears to be strictly developmentally regulated inthe mouse fetal gonads [4]. Both in humans and in mice,Sty is also expressed in the adult testis [1, 2]. In the post-natal mouse testis, the site of Sty expression correspondsto the germ cell rather than to the somatic cell compart-ment, with maximal accumulation in postmeiotic stages [5],i.e., in round spermatids (r.s.). To study Sry function invivo, transgenic mice have been produced carrying thewhole Sy genomic locus. Sry expression in some femaletransgenic mice induced sex reversal [3]. However, the sig-
Accepted June 27, 1997.Received April 25, 1997.'This work was supported by grants from CNR n. 96.031 64.CT04 and
n. 96.04969.ST74, and from MURST 40%.2Correspondence: Pellegrino Rossi, Dipartimento di Sanita' Pubblica
e Biologia Cellulare, Universita' di Roma Tor Vergata, via 0. Raimondo8, 00173, Rome, Italy. FAX: 39-6-72596268; e-mail: [email protected]
nificance of Sry expression in germ cells of the adult testisis unknown.
The peculiar structure of the mouse genomic Sry locusis characterized by two arms of a large inverted repeat,which flank a unique region [6]. Expression studies showedthat Sry RNA is present in somatic cells of genital ridges(g.r.) as a linear molecule of a predicted size of about 5kilobases (kb), whose transcription arises from the uniqueregion of the Sty genomic locus and spans into the 3' armof the repeat [7, 8]. In the unique region, a putative openreading frame (ORF) has been localized that encodes a tran-scription factor containing an HMG box with specific DNAbinding properties [9]. This HMG box is strongly homol-ogous to those of a large family of Sry-related genes, whichhave been termed Sox [10].
In adult germ cells, Sry RNA is a circular molecule ofabout 1.3 kb and is probably generated by the invertedsplicing of a linear RNA precursor containing both the in-verted repeats, which pair and generate a large stem-loopstructure [11]. This inverted splicing event probably in-volves the utilization of a 5' acceptor and a 3' donor splicesite flanking the coding exon, which are located at positions8201 and 9432, respectively, within the unique region ofthe mouse Sy genomic locus. Thus, a circular Sry exon isgenerated, which contains the putative ORF, with a newstop codon derived from the splicing event [ 11 ]. It has beenpostulated that the linear precursor of circular Sry RNAspans several kilobases upstream and downstream of theunique region in the 5' and 3' arms of the inverted repeat[7, 11]. However, in vitro studies indicate that as little as400 base pairs (bp) of the inverted repeat in the linear pre-cursor RNA are required to generate the Sry circular tran-script [12].
The present study was undertaken to map the 5' end ofthe linear precursor of Sry circular RNA and to identify andcharacterize the promoter region that drives its specific ex-pression in male postnatal germ cells. Interestingly, wefound that the Sry circular RNA and its linear precursor arespecifically expressed not only in adult germ cells but alsoin male embryonal gonads, one day after the appearance ofthe linear g.r.-specific transcript that starts from the uniqueregion of the mouse Sty genomic locus.
MATERIALS AND METHODS
Preparation of Biological Materials
Staging and sexing of male g.r. [13], preparation of germcell suspension from adult mouse testis [14], isolation ofr.s. through elutriation techniques [14, 15], and Sertoli cellcultures [16, 17] were performed as previously described.Immediately after isolation, tissues and cells were lysed inRNAzol, and RNA was extracted according to the manu-facturer (Cinna Biotecx Laboratories Int'l, Inc., Friends-wood, TX). Total RNAs were resuspended in diethylpyro-carbonate (DEPC)-treated H20 and used for reverse tran-scription (RT)-polymerase chain reaction (PCR), RNase
1128
Sry PROMOTER ACTIVE IN GERM CELLS AND GENITAL RIDGES
0.5 Kb
P6 PB PA P4 P11 I I I
Unique Reion
W
P5 P2
\l7
P3 B S 0 0 0
P3 B1S1 S2 B2 R1 El\-a\ /--- I
0
8 8P2 P5
<- -
O O
- 0
P1 P4t-a
OC N
PA PB
1./
0
P6I
5' repeat I I F 3' repeat I5' repeat IRF I l 3' repeat
positiveRT-PCRproducts
SA8201
SD9432
Approximate limits of the circular Sry RNA precursor
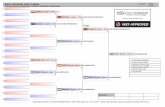
FIG. 1. Schematic representation of the results of RT-PCR analysis performed with total RNA from either 11.5-12.5 dpc male g.r. or r.s., using primersspanning the mouse Sry genomic locus. Each amplification was also performed omitting the reverse transcription step to exclude genomic DNAcontamination. All experiments were performed at least five times, using different RNA preparations, and gave consistent results. Arrows indicate theorientation (forward or reverse) of the primers that we used, whose sequence is reported in Figure 2. SA and SD indicate position of splice acceptorand donor sites utilized for generation of the circular transcript including the ORF encoding an HMG box containing transcription factor [111. Thededuced approximate limits of the linear precursor of the circular form of Sry RNA are indicated on the bottom.
protection assay, and primer extension experiments. Nucle-ar extracts for in vitro transcription were prepared fromfreshly isolated testicular germ cells as previously described[18]. Preparation of nuclear extracts for gel-shift analysisfrom adult germ cells [18] or Sertoli cells [19] has beendescribed previously. Protein concentration (measured withthe Bradford method) of nuclear extracts for both in vitrotranscription and DNA-binding experiments was -10mg/ml.
RT-PCR Analysis
One microgram of total RNA from g.r. and r.s. was re-verse-transcribed in a 2 0-pl reaction in the presence or ab-sence of murine leukemia virus RT (Perkin Elmer, Irvine,CA). Random hexamers or oriented reverse primers wereincluded. PCR amplification was performed in a 50-p1 re-action containing 10 1 l from the reverse transcription stepin the presence of 1 U of Amplitaq (Perkin Elmer) and 0.15F.M primers. Samples were denatured at 95°C for 3 min inthe first cycle and then subjected to 35 step cycles of de-naturation at 95°C (30 sec), annealing at 600C (30 sec), andextension at 72°C (1 min), with a final elongation step of7 min at 72°C. Ten microliters of the reactions were elec-trophoresed on a 1.5% agarose gel and stained by ethidiumbromide.
RNase Protection Assay Experiments
PCR products obtained by amplification of mouse malegenomic DNA with primers P6-PB (-900 bp) and P1-P3(-1300 bp), which are indicated in Figure 1 and describedin Figure 2, were cloned in a PCRII vector (Invitrogen, SanDiego, CA), generating plasmids P6-PB/PCRII and P1-P3/PCRII. In order to reduce the length of the antisense RNAprobe, plasmid P6-PB/PCRII was cut by Sac I (which cutsboth in the vector 5' of the cloning site and within theinsert) and religated, generating plasmid P6-PB/Sac. Onemicrogram of the P6-PB/Sac and P1-P3/PCRII plasmidswere then linearized by HindIII and Acc I digestions, re-spectively, phenol/chloroform-extracted, precipitated, andresuspended in H2 0 for RNA polymerization. A pT7-3-actin antisense control template (Ambion, Austin, TX) wasalso used for riboprobe preparation.
Antisense RNA probes were generated in the presenceof 50 VICi of [a- 3 2P]cytidine triphosphate (CTP; Amersham,Arlington Heights, IL; 800 Ci/mmol), 1.6 I1 of 5-strengthtranscription buffer (Stratagene, La Jolla, CA), 1 1 of 3.3mM each ATP, GTP, and uridine triphosphate (UTP), 0.5
1l of 0.1 mM CTP, 0.5 l1 of 0.2 M dithiothreitol (DTT),1 1l RNasin (60 U/ml, Amersham), and 1 l of T7 RNApolymerase (Stratagene), in a total volume of 8 1. After 1h of incubation, the DNA templates were degraded by theaddition of 1 Il1 of RNase-free DNase I (Stratagene) for 15min. The probes were then purified by denaturing gel elec-trophoresis. Five micrograms of total RNA from 10.5 dayspostcoitum (dpc) g.r., 12.5 dpc g.r., and r.s., and yeast-RNAas a control, were hybridized overnight at 45C in the pres-
P6: CAATTTGCTTGCAACTCTCCTCCT (5362 forward, 12317 reverse)PA: TCCAGTAGTGCCACCTAACTGTTCCATTG (6230 forward, 11449 reverse)PB: TGGAACAGTTAGCTGGCACTACTGGA (6255 reverse, 11424 forward)P4: TCACATTAATCTCCCTCGATGCTTCCC (6585 reverse, 11094 forward)P1: ATTCTTAGCTAGCCCTTGAAAACACAGC (6926 forward, 10753 reverse)P2: TCCCAGTCATTAGAATGCTCAACCTGGATGA (7454 forward, 10246 reverse)P5: TCATCCAGGTTGAGCATTCTAATGACTGGGA (7484 reverse, 10220 forward)P3: AGATATCATGTGGCTGTAGGGCTGAACACT (8250 reverse)B1: TTGTCTAGAGAGCATGGAGGGCCAT (8291 forward)S1: AAGCGCCCCATGAATGCATTTATGGT (8319 forward)S2: ACACTTTAGCCCTCCGATGAGGCTGA (8550 reverse)82: AAAATGCCACTCCTCTGTGACACT (8569 reverse)R1: CACACCTGTACTCCAAAAACCAGCAAAGCT (8665 forward)El: AAATCATAGCAAGGGGGAGTGTTGGCA (9540 reverse).SA/SD: AGCTGACATCACTG/CATTTCAGATCTT (circular Srty RNA-specific, forward)SH1: GTCCTGTATCTATGGCTCTCTGCA (5991, forward)SH2: CCATCCCAATGGGTGCTTATGTCTC (6111, reverse)SH3: GACATAAGCACCCATTGGGATGGA (6089, forward)BM: TGACATGGAGAAACAATAGCAGAA (6049, forward)BM1: GTTTCTGCTATTGTTTCTCCATGTC (6074, reverse)M: TGACATGGAGAGAGTACAGCAGAA (6049 forward, mutated)M1: GTTTCTGCTGTACTCTCTCCATGTC (6074 reverse, mutated)Hprtl: GAAAGACTTGCTCGAGATGTCATG (forward)Hprt2: TGGCAACATCAACAGGACTCCTCG (reverse)
FIG. 2. Oligonucleotide list. Numbers in parentheses indicate positionof the 5' base on the genomic Sry locus; two coordinates are indicatedfor primers hybridizing in both arms of the inverted repeat, and a singlecoordinate is indicated for those hybridizing in the unique region, asshown in Figure 1. SA/SD is the specific primer used for detection ofcircular Sry RNA (the slash indicates the acceptor/donor junction between8201 and 9432, used for splicing out of the circular Sry exon from thelinear precursor RNA). M is identical to BM but contains a 4-nt mutationwithin the core AACAAT motif. M1 is identical to BM1 but contains a4-nt mutation within the core ATTGTT motif. Hprtl and Hrpt2 are primersused for Hprt RNA amplification in RT-PCR experiments (from [51).
1129
DOLCI ET AL.
ence of 3 x 104 cpm of each antisense probe in a hybrid-ization buffer containing 40 mM PIPES [pH 6.7], 80% for-mamide, 0.4 M NaCI, and 1 mM EDTA. The reactions werethen treated with RNase A and T1 (Ambion) for 30 min.After RNase inactivation, the reactions were precipitatedand directly resuspended in loading buffer (Ambion). Theprotected fragments were electrophoresed on a 5% acry-lamide-7 M urea gel and detected through autoradiographyafter 1 wk of exposure at - 800 C.
In Vitro Transcription and Gel-Shift Analysis
Five micrograms of P6-PB/PCRII template were linear-ized by digestion with Pvu I, which cuts twice in the vector,yielding a template containing the P6-PB Sry genomic se-quence flanked by vector sequences upstream and down-stream of the cloning site. After phenol chloroform extrac-tion, two thirds of the digestion product was subsequentlyredigested with Sac I or Xba I. Sac I cuts within the ge-nomic P6-PB sequence and yields an 828-bp template upto the Pvu I site. Xba I cuts both in the P6-PB insert andinside the polylinker downstream from the cloning site,yielding an 810-bp template. A PCR-amplified fragmentcorresponding to the 6089-6585 region of the mouse Srygenomic locus was obtained using primers SH3 and P4 (seeFigure 2). Run-off transcription reactions were performedfor 60 min at 25C, using 10 pxl of nuclear extracts, 0.33-1 1l of a 1 pxg/pxl linearized plasmid DNA template or PCR-amplified product, 10 1l of a buffer containing 14% glyc-erol, 20 mM KC1, 6.5 mM MgCl 2, 14 mM HEPES [pH7.9], 1.2 mM of each ATP, CTP, and UTP, 0.05 mM GTP,0.5 pl1 RNAsin, and 0.5 pl1 [ot- 32 P]GTP (Amersham; 400Ci/mmol, 10 mCi/ml). Reactions were stopped with 380 pl1
of a buffer containing 50 mM Tris-HCl [pH 7.5], 1% SDS,5 mM EDTA, and 25 mg/ml tRNA, extracted twice withphenol-chloroform, precipitated, desiccated, and electro-phoresed on 4% acrylamide gels containing 7 M urea. Forautoradiography, gels were exposed for three days at- 800 C.
PCR fragments for gel-shift assays were directly labeledwith [ot- 32P]deoxy(d) ATP during amplification, whereassynthetic oligonucleotides were filled-in with [- 32 P]dATPand Klenow enzyme (Boehringer, Mannheim, Germany).Labeled PCR fragments were purified by nondenaturingpolyacrylamide gel electrophoresis, whereas labeled oli-gonucleotides were purified by gel filtration on SephadexG50 (Pharmacia, Kalamazoo, MI). Conditions utilized forgel-shift analysis using 5 g of protein have been describedpreviously [18, 19].
RESULTS
RT-PCR Studies
Sry transcript size was studied by RT-PCR analysis, us-ing primers located either in the unique region of the Srylocus or in the inverted repeats (for the oligonucleotide de-scription, see Figure 2). To exclude the possibility thatRT-PCR-generated signals were due to contamination ofRNA samples with genomic DNA, all the experiments werecarried out both in the presence and in the absence of RTin the reverse transcription step of the reactions. Only sig-nals obtained exclusively in the presence of RT were con-sidered to be generated by RNA. To establish which oneof the two inverted repeats was generating positive signals,the reverse transcription step was also performed with ori-ented reverse primers. For instance, the PA-P4 fragment
FIG. 3. The circular form of Sry RNA is expressed in male g.r. between11 .5 and 13.5 dpc, and both linear and circular Sry transcripts are absentin tissues surrounding the male embryonal gonad at 12.5 dpc. A) RT-PCRanalysis of total RNA from male g.r. at the indicated time of differentia-tion, using B2 as reverse primer, and either B1 or SA/SD as forward prim-er. All samples were also amplified with Hprt primers as a control forRNA loading. No products were detectable on omitting RT from the re-action (these reactions were loaded in the same lanes together with HprtRT-PCR products). In the first lane, DNA molecular standards (nt) VI(Boehringer) were loaded. B) RT-PCR analysis of total RNA from the bodywall of 12.5 dpc embryos, using the same primers indicated in A. Resultsof reactions with Sry primers performed both in the presence (+) and inthe absence (-) of RT shown. In the second lane, 1 Kb DNA molecularweight markers (Gibco BRL, Grand Island, NY) were loaded. B1-B2 am-plify 278 bp from both linear and circular Sry RNA molecules, whereasSA/SD-B2 amplify 382 bp only from the circular form, since SA/SD over-laps the acceptor/donor junction generated after splicing out of the cir-cular exon [1 1, as schematically explained on the right.
was assigned to the left arm of the repeat if the reversetranscription step was performed with primer P4, or to theright arm if the reverse transcription step was performedwith primer PA (Fig. 1). Using total RNA either from 11.5-12.5 dpc male g.r., or from r.s., positive RT-PCR signalswere detected up to position 6230 of the published se-quence [6] of the Sty genomic locus (Fig. 1). No RT-PCRsignals were detected with more upstream primers (for in-stance P6 in Fig. 1), which were able to amplify genomicDNA. This suggested that the transcriptional initiation siteof the linear precursor of the circular form of Sr' RNA islocated in the 5' arm of the inverted repeat between 5362and 6230. The presence of RNAs extending in the 5' armof the inverted repeat not only in r.s. but also in 11.5-12.5dpc male g.r. suggested that the circular Sry RNA mole-cules might be present also in the male embryonal gonad.Indeed, by using as forward primer SA/SD, which can de-tect only the circular form of Sry RNA [11], we identifiedthe predicted RT-PCR signal in male g.r. as early as 11.5dpc. and up to 13.5 dpc (Fig. 3A), but not in the surround-ing tissues (Fig. 3B).
As for the 3' end of Sry transcripts, our RT-PCR analysisindicated that it should map between positions 11449 and12317 (Fig. 1). This could correspond either to the 3' endof the linear precursor of circular Sty RNA or to the 3' endof the linear form of the transcript starting within the
1130
Sry PROMOTER ACTIVE IN GERM CELLS AND GENITAL RIDGES
unique region, which has indeed been reported to terminateat position 11575 in g.r. [8].
RNase Mapping Studies
Mapping of the 5' end of the Sty circular RNA precursorin both g.r. and r.s. was performed by RNase protectionassay. An antisense 32 P-end-labeled RNA molecule, corre-sponding to the 5640-6255 region (probe A) protected twodiscrete RNA regions of -80 and -60 nucleotides (nt) in12.5 dpc g.r. and in r.s., but not in 10.5 dpc g.r. (Fig. 4A).Thus, the origin of the linear precursor of the circular Srtytranscript should be around positions 6170 and 6190, in the5' arm of the inverted repeat, both in r.s. and in 12.5 dpcg.r., and this transcript should be absent in 10.5 dpc g.r. Thesamples contained similar RNA amounts, as indicated by aparallel RNase protection assay with a 3-actin riboprobe(Fig. 4B). The same RNA preparation from 10.5 dpc g.r.,used in RNase protection assay experiments with an anti-sense probe spanning the unique region (7660-8250, probeB), yielded the predicted band of -230 nt (Fig. 4C), con-firming that in g.r. a linear transcript originates from a pro-moter within the unique region [7, 8] as early as at 10.5dpc, when the linear precursor of the circular Sry transcriptis not yet detectable. Indeed, no full protection of antisenseprobe B was evident with RNA from 10.5 dpc g.r., evenafter much longer exposure times. The -230-nt band pro-tected by antisense probe B was not detected in RNA prep-arations from r.s. (data not shown), confirming that lineartranscripts originating within the unique region are absent(or, at least undetectable) in adult germ cells [7].
Computer Analysis
The possibility that the protection assay might be mask-ing intron-exon boundaries around positions 6170 and 6190of the Sry genomic locus could not be excluded, since prim-er extension with RNA from adult germ cells using an end-labeled primer starting from position 6255 and 5' rapid am-plification of cDNA ends (RACE)-PCR experiments wereinconclusive (data not shown). However computer analysisof the Sry genomic locus (performed with programs fromPC/GENE, 6.85 release [Intelligenetics, Mountain View,CA] on the sequence from ref. [6]; EMBL accession num-ber: X67204) revealed the absence of consensus acceptorsplice sites in this area. Computer analysis also showed thattwo potential transcriptional initiation sites for the Sry cir-cular RNA precursor actually map at positions 6178 and6197, 24 and 43 bp downstream from a TATA sequencecentered at 6154, and 72 and 91 bp downstream from aninverted CCAAT box starting at 6106, strongly suggestingthat a transcriptional promoter might be present in this areaof the 5' arm of the inverted repeat (see Fig. 6, bottom).This area of the mouse Sry genomic locus shares a limitedhomology with a region of the human SRY genomic locus,which is located at a similar distance upstream from theORF encoding the transcription factor [7]. Interestingly, inclose proximity upstream to the two potential transcription-al initiation sites in the mouse Sry genomic locus, a se-quence corresponding to the consensus high-affinity Srybinding site AACAAT [20] is found between 6060 and6065, and its complement ATTGTT between 6169 and6174 (see Fig. 6, bottom).
Since identical sequences are present in the 3' arm ofthe inverted repeat, the possibility exists that RT-PCR anal-ysis with oriented reverse primers (Fig. 1) and RNase pro-tection assay with probe A (Fig. 4A) were detecting anti-
FIG. 4. Two upstream ends of the linear precursor of the circular formof Sry RNA in male g.r. and adult germ cells map -80 and -60 nt up-stream from position 6255 of the mouse Srygenomic locus. A) Represen-tative RNase protection assay experiment using an antisense riboprobe(probe A) spanning nt 5640-6255 of the Sry genomic locus with totalRNAs from 10.5 and 12.5 dpc g.r., and from r.s. This experiment wasrepeated three times, with identical results. B) As controls for RNA load-ing, the same RNA sources as in A were subjected to RNase protectionassay using an antisense ,-actin riboprobe. C) RNase protection assaywith the same RNA sample from 10.5 dpc g.r. used in A, utilizing anantisense riboprobe (probe B) spanning nucleotides 7660-8250 of the Srygenomic locus. Molecular sizes were determined according to the mo-bility of end-labeled DNA molecular standards (nt) VI (Boehringer). Aschematic representation of RNase mapping experiments is drawn on thebottom.
1131
DOLCI ET AL.
FIG. 5. A transcriptional promoter between positions 5640 and 6197in the 5' arm of the inverted repeat of the mouse Sry genomic locus isfunctional in vitro in adult germ cells. A) Autoradiography of labeledRNAs produced in run-off transcription experiments with nuclear ex-tracts from a mixture of meiotic and postmeiotic germ cells, using astemplates plasmid P6-PB/PCRII (see Materials and Methods) linearizedand/or deleted with the indicated restriction enzymes. Plasmid digestedwith Pvu I (0.33 g) and 1 pLg of the same plasmid digested with PvuI/Sac I or Pvu IlXba I were used in the reactions. Arrows indicate bandswhose size matches that predicted for a transcription starting from po-sition 6197 or 61 78 of the Sry genomic locus. The upper band representsan aspecific transcript, since it was observed with all templates. A sche-matic representation of the results of the experiment is shown on theleft. B) Autoradiography of labeled RNAs produced in run-off transcrip-tion experiments with nuclear extracts from a mixture of meiotic andpostmeiotic germ cells, using as template 1 -g of a PCR-amplified DNAfragment (obtained with primers SH3 and P4) corresponding to the6089-6585 region of the mouse Sry genomic locus. A schematic rep-resentation of the results of the experiment is shown on the bottom.CAP, transcriptional initiation sites. Molecular sizes were determinedaccording to the mobility of end-labeled DNA molecular standards (nt)VI (Boehringer).
sense transcripts arising from the 3' arm. In order to verifywhether a potential promoter in the symmetrical region ofthe 3' arm drives the expression of antisense Sy RNAs,we used as oriented reverse primers from the unique regionoligonucleotides B1, S1, and R1 (corresponding to forwardprimers of the sense strand) in the reverse transcription stepof RT-PCR experiments, followed by PCR amplificationusing as forward primers oligonucleotides B2, S2, and E1l(corresponding to reverse primers of the sense strand). Withthis study, we did not obtain any evidence for the expres-sion of antisense STy transcripts either in g.r. and in r.s. (datanot shown). Thus, this potential promoter is active only inthe 5' arm of the inverted repeat.
In Vitro Transcription Studies
The presence of a transcriptional promoter upstream toposition 6178/6197 was confirmed in vitro, by performingrun-off transcription experiments using the 5362-6255 re-gion of the Sty locus as a template and nuclear extractsfrom a mixture of meiotic and postmeiotic germ cells from
adult testis. Three types of restriction digestions (Pvu I, PvuI/Sac I, and Pvu IXba I) were performed on the templatein order to monitor the length of the transcription products.A major transcript of the predicted size (starting from 6197)was generated with all templates (Fig. 5A; note that a 3-foldlower concentration of DNA template was used in the PvuI reaction shown in the first lane), and a minor transcript(starting from 6178) was also evident with the Pvu IlXba Idigestion (Fig. 5A, lane 3). Since one of the restrictionenzymes that we used to linearize the template (Sac I) cutsalso at position 5640 (Fig. 5A, second lane), we can con-clude that a promoter element active in nuclear extracts ofadult germ cells is present between 5640 and 6178/6197.
To define the minimal promoter region, we also per-formed in vitro transcription experiments using a PCR-am-plified genomic region between 6089 and 6585 (obtainedwith primers SH3 and P4) with germ cell nuclear extracts.This fragment lacks the Srv consensus binding site AA-CAAT present between 6060 and 6065 but still contains itscomplement ATTGTT between 6169 and 6174, togetherwith both the inverted CCAAT box and the TATA boxsequences (see Fig. 6, bottom). As shown in Figure 5B,two RNA species of the expected size (approximately 390and 410 nt) were still transcribed from this template, eventhough the intensity of these bands was weak, if comparedto the intensity of specific bands obtained with the sameconcentration of a template spanning the 5640-6255 region(see the Pvu IVSac I lane in Fig. 5A). These data furtherconfirm that two transcriptional initiation sites utilized ingerm cells are located at 6178 and 6197. They also indicatethat a minimal promoter active in germ cells is located be-tween 6089 and 6178/6197, even though a stronger tran-scriptional activity is obtained when sequences between5640 and 6089 are also present.
DNA-Binding Studies
To verify whether specific transcription factors bind tothe regulatory sequences upstream to the transcriptionalstart sites, gel mobility shift assay experiments were per-formed using nuclear extracts from adult germ cells. TwoPCR-amplified fragments from position 5991 to 6111 (ob-tained with primers SHI and SH2) and from 6089 to 6255(obtained with primers SH3 and PB) were radiolabeled andincubated with nuclear extracts from freshly isolated adultgerm cells or from Sertoli cells.
With 5991-6111 used as a probe, a retarded band wasobserved in nuclear extracts from germ cells but not fromSertoli cells (Fig. 6, left panel). A similar electrophoreticpattern was observed when we used 6089-6255 as a probe(Fig. 6, middle panel). Binding specificity was demonstrat-ed by a complete competition of the DNA-protein com-plexes by the addition of a 250-fold molar excess of thesame unlabeled sequences. A complete inhibition of theDNA-protein complex formed with the 6089-6255 frag-ment was obtained when 5991-6111 was used as a com-petitor (Fig. 6, middle panel), but not after the addition ofthe same molar excess of an unrelated sequence (the Sryexonic region between 8319 and 8550, obtained with prim-ers S1 and S2), suggesting that the nucleotide sequencecontained in 6089-6255 is recognized by the same nuclearproteins binding the 5991-6111 fragment.
As mentioned in the previous paragraphs, a sequencecorresponding to the consensus high-affinity Sry bindingsite AACAAT [20] is found between 6060 and 6065 in the5991-6111 fragment, and its complement ATTGTT be-
1132
Sry PROMOTER ACTIVE IN GERM CELLS AND GENITAL RIDGES
FIG. 6. Two high-affinity Sry binding sites [201 in the proximal promoter in the 5' arm of the inverted repeat of the mouse Sry genomic locusrecognized a nuclear factor present in adult germ cells but not in Sertoli cells. Left) Gel-shift analysis using nuclear extracts from a mixture ofmeiotic and postmeiotic g.c. or from Sertoli cells, using as a probe a PCR-amplified fragment (obtained with primers SH1 and SH2) spanningcoordinates 5991-6111 of the mouse Sry genomic locus. The same unlabeled fragment was used as a competitor (250-fold molar excess, last lane).Middle) Same experiment using a PCR-amplified fragment (obtained with primers SH3 and PB) spanning coordinates 6089-6255 of the mouse Srygenomic locus. Either the same fragment or fragments spanning 5991-6111 or 8319-8550 (obtained using primers S1 and S2) of the locus wereused as unlabeled competitors (250-fold molar excess). Right) Same experiment using a double-stranded synthetic oligonucleotide (oligonucleotidesBM and BM1) spanning coordinates 6049-6072 of the mouse Sry genomic locus. Competition was performed both with the 6049-6072 double-stranded oligonucleotide (w.t.) or a double-stranded oligonucleotide (oligonucleotides M and M1) in which the high-affinity Sry binding site (AA-CAAT) was mutated to GAGTAC (mut.) at the indicated molar excess. The sequence between 5991 and 6200 of the mouse Sry genomic locus fromref. [6] (corresponding to the proximal promoter region from which the linear precursor of circular Sry transcript is generated) is reported on thebottom. The two high-affinity Sry binding sites are boxed. An inverted CCAAT box and the TATA box are underlined. Arrows indicate the two majortranscriptional initiation sites.
tween 6169 and 6174 in the 6089-6255 fragment, sug-gesting that the nuclear proteins binding to both fragmentsmight recognize this sequence motif. To verify that theseAACAAT motifs might be involved in the formation ofDNA-protein complexes, we used a synthetic 24-bp double-stranded oligonucleotide spanning 6049 to 6072 (containingthe AACAAT consensus sequence, and obtained with prim-ers BM and BM1) and a 24-bp double-stranded oligonu-cleotide with a 4-bp mutation within the consensus (ob-tained with primers M and Ml). Using radiolabeled 6049-6072, a retarded band was observed with nuclear extractsfrom germ cells but not from Sertoli cells (Fig. 6, right
panel). Competition by a 50- and a 100-fold molar excessof cold 6049-6072 completely abolished formation of thisDNA-protein complex, whereas incubation with the samemolar excess of the double-stranded oligonucleotide withthe 4-bp mutation was much less effective for competition.
DISCUSSION
The major aim of the present study was the identificationof the regulatory regions that drive the expression of thelinear precursors of the circular Sry transcript in the adultmouse testis. By using RT-PCR and RNase protection as-
1133
DOLCI ET AL.
say, we showed that these RNA molecules originate in the5' arm of the inverted repeat starting from position 6178and 6197 of the Sry genomic locus, both in the male em-bryonal gonad and in adult germ cells. In vitro transcriptionexperiments using nuclear extracts from adult germ cellsindicate that authentic transcriptional initiation sites map at6178 and 6197, and that a minimal promoter region activein these cells is present between 6089 and 6178/6197 (Fig.5). These results support the evidence by computer analysisthat no consensus splice acceptor sites are present in theSry genomic locus around the 5' limit of protection withprobe A in our RNase mapping experiments. Others haveshown full RNase protection in the adult testis with an an-tisense riboprobe spanning the 5046-5490 region [7], butwe did not detect any full protection upstream of 6178 withthe 5640-6255 antisense riboprobe (probe A, Fig. 4A),raising the possibility that splicing events utilizing an ac-ceptor site in the area between 5490 and 5640 did not letus detect transcripts generated from other promoters lyingfurther upstream in the 5' arm of the inverted repeat.
Interestingly, two consensus high-affinity Sy-bindingsites are found between 6060 and 6065 and between 6169and 6174. These sequences are contained within fragments5911-6111 and 6089-6255, which are specifically recog-nized in gel-shift analysis by an identical protein factorpresent in nuclear extracts from adult germ cells but notfrom Sertoli cells. Furthermore, a double-stranded oligo-nucleotide spanning the 6049-6072 area, but not an oli-gonucleotide containing a 4-bp mutation within the AA-CAAT consensus found between 6060 and 6065, also rec-ognizes nuclear factors present in germ cells but not inSertoli cells. These data strongly suggest that transcriptionfrom this promoter in mouse germ cells might be regulatedby a transcription factor that recognizes these two AA-CAAT motifs. This transcription factor might be some othermember of the Sox family expressed in the postnatal testis[21], if not the Sry transcription factor itself. The impor-tance of the AACAAT motif present between 6060 and6065 is further suggested by the observation that the 6089-6585 template lacking this sequence is much less efficientin run-off transcription experiments with respect to the5640-6255 template.
No data are available on the involvement of Sty in adulttesticular function, and the significance of circular Sry tran-scripts in adult mouse germ cells is unknown. The inter-pretation that these transcripts in the adult testis are non-functional, being scarcely associated to polyribosomes [ 11],is not conclusive, since circular RNAs can be translated ineukaryotic systems, provided that they contain internal ri-bosomal entry sites [22], and mRNAs that are translated inlate spermiogenesis (such as that for protamine) are notassociated with polysomes in r.s. [23]. Moreover, translationof the Sry protein from the linear precursor RNA, insteadof from the circular splicing product, cannot be excluded.
Our present study shows that the circular Sty RNA andits precursor are present not only in germ cells from thepostnatal testis, but also in male g.r. The latter finding isonly apparently in conflict with previous reports [7, 11].Indeed, Capel et al. [ 11] concluded through RT-PCR anal-ysis that circular Sry transcripts are rare in male embryonalgonads, but Hacker et al. [7] have shown by RNase map-ping experiments that a substantial proportion of the Stytranscripts at 11.5 dpc g.r. are generated by utilization ofthe splice donor in the unique region at position 9432,which is utilized in adult germ cells to generate circular Srytranscripts. This is consistent with our RT-PCR evidence
for the presence of circular Sry transcripts in male g.r. start-ing at 11.5 and up to 13.5 dpc, and with our evidencethrough both RT-PCR and RNase mapping that the linearRNA precursor is present in g.r. at 12.5 dpc. RT-PCR anal-ysis previously performed with primers that do not discrim-inate between linear and circular transcripts [4, 5] suggeststhat Sry circular transcripts and their precursors, as well aslinear transcripts starting from the unique region, are allexpressed in somatic cells of male g.r. rather than in pri-mordial germ cells.
The two consensus high-affinity Sry binding sites pres-ent in the promoter that we have mapped in the 5' arm ofthe inverted repeat are suggestive of a regulatory role inmale g.r. for the Sry protein probably translated from thelinear transcript originating within the unique region at10.5 dpc [7, 8]. Indeed, according to the present data, thecircular transcript is detectable in the male embryonal go-nad at 11.5 dpc, i.e., one day after the expression of theunique region-specific linear transcript, and up to 13.5dpc. The expression of the circular Sry RNA in male g.r.and of its linear precursor, unlike that of the unique region-specific linear transcript, precedes closely the reported ex-pression of transcripts from other genes involved in thecascade of testicular differentiation, such as the one en-coding the anti-Miillerian hormone [7]. We have previ-ously shown that expression of the Xist gene, a marker ofX-chromosome inactivation, occurs not only during sper-matogenesis, but also in somatic cells of male g.r. between11.5 and 13.5 dpc [13]. Recently, X-chromosome inacti-vation, which normally occurs in all somatic cells in fe-males to ensure dosage compensation, but also duringspermatogenesis in males, has been shown actually to oc-cur also in somatic cells of male embryonal gonads, asevaluated through lack of expression of an X-linked trans-gene [24]. Since expression of circular Sry transcripts andexpression of the Xist gene with consequent X-chromo-some inactivation are parallel phenomena during bothmale sex determination and spermatogenesis, the questionarises whether the Sy gene, through either its linear or itscircular RNA products, might play a role in the control ofX-chromosome inactivation in males. X-inactivation is es-sential for spermatogenesis after birth [25], but it could bealso involved in sex determination, as suggested by thediscovery of a dosage-sensitive locus on the short arm ofthe human X-chromosome, which, when duplicated, caus-es male-to-female sex reversal [26].
The present identification of a promoter region gener-ating the precursor of the Sry circular RNA, and possiblyregulated by the Svty transcription factor itself or by theproducts of other Sox genes, should help to establish thefunction of this peculiar transcript in testicular differenti-ation in the mouse embryo, and its eventual role in sper-matogenesis after birth. This could be achieved by com-paring the effect of genetic ablation through homologousrecombination of the promoter sequences that we haveidentified in the 5' arm of the inverted repeat (which gen-erate the precursors of circular transcripts) with the effectof ablation of the as yet unidentified promoter sequencesthat should lie within the unique region of the Sry genomiclocus (which generate conventional linear transcripts).
ACKNOWLEDGMENT
We thank Dr. Cristina Albanesi for her help in preparation of nuclearextracts for in vitro transcription experiments.
1134
Sry PROMOTER ACTIVE IN GERM CELLS AND GENITAL RIDGES
REFERENCES
1. Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Min-sterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mappingto the sex-determining region of the mouse Y chromosome is a mem-ber of a novel family of embryonically expressed genes. Nature 1990;346:245-250.
2. Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, SmithMJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. Agene from the human sex-determining region encodes a protein withhomology to a conserved DNA-binding motif. Nature 1990; 346:240-244.
3. Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R.Male development of chromosomally female mice transgenic for Sry.Nature 1991; 351:117-121.
4. Koopman P, Miinsterberg A, Capel B, Vivian N, Lovell-Badge R.Expression of a candidate sex-determining gene during mouse testisdifferentiation. Nature 1990; 348:450-452.
5. Rossi P, Dolci S, Albanesi C, Grimaldi P, Geremia R. Direct evidencethat the mouse sex-determining gene Sry is expressed in the somaticcells of male fetal gonads and in the germ cell line in the adult testis.Mol Reprod Dev 1993; 34:369-373.
6. Gubbay J, Vivian N, Economou A, Jackson D, Goodfellow P, Lovell-Badge R. Inverted repeat structure of the Sry locus in mice. Proc NatlAcad Sci USA 1992; 89:7593-7597.
7. Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Expression ofSry, the mouse sex-determining gene. Development 1995; 121:1603-1614.
8. Jeske YWA, Bowles J, Greenfield A, Koopman P. Expression of alinear Srv transcript in the mouse genital ridge. Nat Genet 1995; 10:480-542.
9. Dubin RA, Ostrer H. Sty is a transcriptional activator. Mol Endocrinol1994; 8:1182-1192.
10. Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, NorrisD, Rastan S, Stevanovic M, Goodfellow PN, Lovell-Badge R. A com-parison of the properties of Sox-3 with Sry and two related genes,Sox-1 and Sox-2. Development 1996; 122:509-520.
11. Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Good-fellow P, Lovell-Badge R. Circular transcripts of the testis determininggene Srv in adult mouse testis. Cell 1993; 73:1019-1030.
12. Dubin RA, Kazmi MA, Ostrer H. Inverted repeats are necessary for cir-cularization of the mouse testis Sry transcript. Gene 1995; 167:245-248.
13. Dolci S, Geremia R, Albanesi C, Rossi P. Expression of the Xist gene
in urogenital ridges of midgestation male embryos. Biochem BiophysRes Commun 1994; 205:334-340.
14. Sorrentino V, McKinney MD, Giorgi M, Geremia R, Fleissner E. Ex-pression of cellular protooncogenes in the mouse male germ cell line:a distinctive 2.4-kilobase pim-l transcript is expressed in haploid post-meiotic cells. Proc Natl Acad Sci USA 1988; 85:2191-2195.
15. Sorrentino V, Giorgi M, Geremia R, Besmer P, Rossi P Expression ofthe c-kit proto-oncogene in the murine male germ cells. Oncogene1991; 6:149-151.
16. Rossi P, Dolci S, Albanesi C, Grimaldi P, Ricca R, Geremia R. Fol-licle-stimulating hormone induction of steel factor (SLF) mRNA inmouse Sertoli cells and stimulation of DNA synthesis in spermato-gonia by soluble SLE Dev Biol 1993; 155:68-74.
17. Rossi P, Grimaldi P, Blasi F, Geremia R, Verde P. Follicle-stimulatinghormone and cyclic AMP induce transcription from the human uro-kinase promoter in primary cultures of mouse Sertoli cells. Mol En-docrinol 1990; 4:940-946.
18. Albanesi C, Geremia R, Giorgio M, Dolci S, Sette C, Rossi P. A cell-and developmental stage-specific promoter drives the expression of atruncated c-kit protein during mouse spermatid elongation. Develop-ment 1996; 122:1291-1302.
19. Grimaldi P, Piscitelli D, Albanesi C, Blasi F, Geremia R, Rossi P.Identification of 3',5'-cyclic adenosine monophosphate-inducible nu-clear factors binding to the human urokinase promoter in mouse Ser-toli cells. Mol Endocrinol 1993; 7:1217-1225.
20. Harley VR, Lovell-Badge R, Goodfellow PN. Definition of a consen-sus DNA binding site for SRY. Nucleic Acids Res 1994; 22:1500-1501.
21. Denny P, Swift S, Connor F, Ashworth A. An Srty-related gene ex-pressed during spermatogenesis in the mouse encodes a sequence-specific DNA-binding protein. EMBO J 1992; 11:3705-3712.
22. Chen C-Y, Sarnow P. Initiation of protein synthesis by the eukaryotictranslational apparatus on circular RNAs. Science 1995; 268:415-417.
23. Kleene KC. Patterns of translational regulation in the mammalian tes-tis. Mol Reprod Dev 1996; 43:268-281.
24. Jamieson RV, Zhou SX, Tan SS, Tam PP. X-chromosome inactivationduring the development of the male urogenital ridge of the mouse. IntJ Dev Biol 1997; 41:49-55.
25. Lifschytz E, Lindsley DL. The role of X-chromosome inactivationduring spermatogenesis. Proc Natl Acad Sci USA 1972; 69:182-186.
26. Bardoni B, Zanaria E, Guioli S, Floridia G, Worley KC, Tonini G,Ferrante E, Chiumello G, McCabe ERB, Fraccaro M, Zuffardi O,Camerino G. A dosage sensitive locus at chromosome Xp21 is in-volved in male to female sex reversal. Nat Genet 1994; 77:497-501.
1135