Genotype-phenotype correlation of contiguous gene deletions ...
PCR-Based Method To Differentiate the Subspecies of the Mycobacterium tuberculosis Complex on the...
-
Upload
independent -
Category
Documents
-
view
3 -
download
0
Transcript of PCR-Based Method To Differentiate the Subspecies of the Mycobacterium tuberculosis Complex on the...
JOURNAL OF CLINICAL MICROBIOLOGY, Apr. 2003, p. 1637–1650 Vol. 41, No. 40095-1137/03/$08.00�0 DOI: 10.1128/JCM.41.4.1637–1650.2003Copyright © 2003, American Society for Microbiology. All Rights Reserved.
PCR-Based Method To Differentiate the Subspecies of theMycobacterium tuberculosis Complex on the Basis of
Genomic DeletionsRichard C. Huard,1,2 Luiz Claudio de Oliveira Lazzarini,1 W. Ray Butler,3
Dick van Soolingen,4 and John L. Ho1*Division of International Medicine and Infectious Diseases, Department of Medicine, Joan and Sanford I. Weill
Medical College1 and Graduate School of Medical Sciences,2 Cornell University, New York, New York;Division of AIDS, STD and TB Laboratory Research, National Center for Infectious Diseases,
Centers for Disease Control and Prevention, Atlanta, Georgia3; and National Instituteof Public Health and the Environment, Bilthoven, The Netherlands4
Received 13 November 2002/Returned for modification 8 January 2003/Accepted 18 January 2003
The classical Mycobacterium tuberculosis complex (MtbC) subspecies include Mycobacterium tuberculosis,Mycobacterium africanum (subtypes I and II), Mycobacterium bovis (along with the attenuated M. bovis bacillusCalmette-Guerin [BCG]), and Mycobacterium microti; increasingly recognized MtbC groupings include Myco-bacterium bovis subsp. caprae and “Mycobacterium tuberculosis subsp. canettii.” Previous investigations havedocumented each MtbC subspecies as a source of animal and/or human tuberculosis. However, study of theseorganisms is hindered by the lack of a single protocol that quickly and easily differentiates all of the MtbCgroupings. Towards this end we have developed a rapid, simple, and reliable PCR-based MtbC typing methodthat makes use of MtbC chromosomal region-of-difference deletion loci. Here, seven primer pairs (whichamplify within the loci 16S rRNA, Rv0577, IS1561�, Rv1510, Rv1970, Rv3877/8, and Rv3120) were run inseparate but simultaneous reactions. Each primer pair either specifically amplified a DNA fragment of aunique size or failed, depending upon the source mycobacterial DNA. The pattern of amplification productsfrom all of the reactions, visualized by agarose gel electrophoresis, allowed immediate identification either asMtbC composed of M. tuberculosis (or M. africanum subtype II), M. africanum subtype I, M. bovis, M. bovis BCG,M. caprae, M. microti, or “M. canettii” or as a Mycobacterium other than MtbC (MOTT). This MtbC PCR typingpanel provides an advanced approach to determine the subspecies of MtbC isolates and to differentiate themfrom clinically important MOTT species. It has proven beneficial in the management of Mycobacteriumcollections and may be applied for practical clinical and epidemiological use.
Mycobacteria that cause tuberculosis in mammals form theMycobacterium tuberculosis complex (MtbC) and include My-cobacterium tuberculosis, Mycobacterium africanum (dividedinto subtype I [group A/West African/“M. bovis-like”] and sub-type II [group B/East African/“M. tuberculosis-like”]), Myco-bacterium bovis (along with the M. bovis-derived bacillusCalmette-Guerin [BCG] vaccine strains), Mycobacterium mi-croti, Mycobacterium bovis subsp. caprae (M. caprae), and “My-cobacterium tuberculosis subsp. canettii” (“M. canettii”) (7, 29)(the name “M. canettii” is in quotation marks since it does notappear on the official List of Bacterial Names with Standing inNomenclature [http://www.bacterio.cict.fr]). Members of theMtbC are highly related mycobacteria exhibiting remarkablenucleotide sequence level homogeneity despite varying inpathogenicity, geographic range, certain physiological features(such as colony morphology as well as profiles of resistance andsusceptibility to inhibitors), epidemiology, and host preference(10, 11, 39). Notably, M. bovis has a wide host range but isprimarily a bovid pathogen, goats are the natural host of M.
caprae, and M. microti is most often isolated from small ro-dents, while M. tuberculosis is the predominant cause of humantuberculosis (2, 3, 39, 44, 45). However, each of the MtbCsubspecies is known to infect humans (16, 17, 20, 22, 27, 33,43–45), and since most laboratories do not fully identify MtbCisolates, the true cause of tuberculosis in these patients and itssource often remain undiscovered. An important health con-cern is the zoonotic transmission of some MtbC subspeciesfrom animals to humans and vice versa. Of particular signifi-cance is the transmission of M. bovis to humans from cattle andunpasteurized milk as well as M. bovis BCG infection of im-munocompromised individuals (1, 21, 27, 33). M. bovis is nat-urally resistant to pyrazidamide, a first-line antituberculosisdrug (31, 36). Therefore, complete identification of MtbC iso-lates at the subspecies level is required in order to collectinformation on their epidemiology and also to enable appro-priate patient treatment and public health measures.
Various biological and molecular mycobacterial characteris-tics have been utilized to identify MtbC isolates but have lim-ited applicability as MtbC taxonomical tools. Although certainMycobacterium species-specific gene sequence differences workwell to differentiate mycobacteria other than MtbC (MOTT)from each other and from the MtbC, to date none can discrim-inate the individual MtbC subspecies due to genetic invariancein the target loci (8, 11, 26, 28, 34, 37, 39, 41). In contrast, a
* Corresponding author. Mailing address: Cornell University, Joanand Sanford I. Weill Medical College, Department of Medicine, Di-vision of International Medicine and Infectious Diseases, RoomA-421, 525 East 68th St., New York, NY 10021. Phone: (212) 746-6316.Fax: (212) 746-8675. E-mail: [email protected].
1637
series of classical tests based upon growth, phenotypic, andbiochemical properties have been traditionally used to segre-gate members of the MtbC (17, 30). However, together thesetests can be slow, cumbersome, imprecise, nonreproducible,and time-consuming, and they may not give an unambiguousresult in every case and may not be performed by every labo-ratory. To complement the classical tests for determination ofMtbC species, well-defined MtbC lineage- and subspecies-re-stricted single-nucleotide polymorphisms (SNPs) in the gyrA,katG, pncA, oxyR�, hsp65, and gyrB genes have been used tospecify certain MtbC groupings through sequence analysisand/or digestion of PCR products followed by restriction frag-ment length polymorphism (PCR-RFLP) analysis (12, 13, 18,30, 31, 36, 38, 39). However, these loci on their own are unableto differentiate all of the MtbC subspecies. Likewise, moleculargenetic MtbC typing assays (e.g., variable numbers of tandemrepeat analysis, mixed linker PCR, and IS6110 RFLP) thathave been designed to reveal interstrain relationships (12, 17,25, 39) cannot be used as efficient taxonomic tools to unam-biguously classify individual MtbC strains. Spacer oligonucle-otide typing (spoligotyping) is the only DNA-based methodol-ogy for which most MtbC members are believed to havesignature features (3, 16, 23, 25, 31, 42–45). However, spoligo-types, the numerical output of spoligotyping, are not necessar-ily exclusive to one MtbC member, nor are they restricted, asstrains can waver from the expected minimal consensus spoli-gopattern for their MtbC subspecies. Hence, an improved pro-tocol for MtbC species determination with greater discrimina-tory power is needed.
Comparative genomics studies employing several differentgenetic hybridization strategies revealed regions of difference(RD) representing the loss of genetic material in M. bovis BCGcompared to M. tuberculosis H37Rv (4, 6, 14). One of thesedeletions is believed to have been the primary attenuationevent in the derivation of M. bovis BCG from M. bovis, since allM. bovis BCG isolates bear this deletion (4). PCR analysis forthese long sequence polymorphisms (LSPs) found some RDloci to be restricted to one MtbC strain or subspecies, whileothers appeared to be differentially distributed among theMtbC groupings (7, 14, 29). These data suggested a sequentialaccumulation of LSPs and was used to construct a phylogeneticmap for the evolution of the MtbC (7). Recently, PCR analysisof certain LSPs, in combination with phenotypic testing, wasshown to accurately differentiate several MtbC groupings andwas used to evaluate an large collection of clinical isolates (35).Unfortunately, some of the targeted RD loci do not haverestricted profiles for particular MtbC subspecies, nor was theprotocol evaluated for the identification of M. caprae, “M.canettii,” and MOTT isolates.
In this study we investigated the RD profile of the MtbCgroupings, and based upon this information we developed asolely PCR-based system using seven PCR primer pairs spe-cific to the loci 16S rRNA, Rv0577, IS1561�, Rv1510, Rv1970,Rv3877/8, and Rv3120, which together form a MtbC PCRtyping panel (Table 1). The final pattern of amplification prod-ucts of all reactions, given by failure or success, clearly segre-gated the tested strains from MOTT isolates and by MtbCsubspecies identity.
(This study contributed to the fulfillment of the Ph.D. de-gree requirements by R. C. Huard.)
MATERIALS AND METHODS
Strains analyzed. A total of 71 MtbC strains and 44 MOTT isolates weretested (Table 2). These strains were characterized by various typing methods, i.e.,numerical analysis of phenotypic and biochemical characteristics, 16S rRNAsequencing, IS6110-RFLP analysis, and spoligotyping. The country of origin ofthese strains (where available), as well as references to prior publications thatutilized certain strains, is given in Table 2. A list associating mycobacterial strainswith the contributing laboratory is available upon request.
DNA preparation for PCR. DNA from mycobacterial cultures was purified asfollows. Cultured bacteria were spun down, resuspended in 1 ml of Tris-EDTAbuffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA), and transferred to a 2-mlEppendorf tube. Lysozyme solution (100 �l; 10 mg/ml in Tris-EDTA buffer) andfive 3-mm-diameter glass beads were then added, vortexed, sonicated for 10 min,vortexed again, and then incubated at 37°C for 2 h with brief vortexing every 30min. To the resultant suspension, after water bath sonication for 10 min anddivision into two 1.5-ml Eppendorf tubes, was added 70 �l of 10% sodiumdodecyl sulfate and 10 �l of proteinase K (10 mg/ml). The mixture was thenvortexed and incubated for 2 h at 65°C, with brief vortexing every 30 min.Afterwards, 100 �l of 5 M NaCl was added and vortexed, and following theaddition of 80 �l of 10% hexadecyltrimethyl ammonium bromide (Sigma, St.Louis, Mo.) in pure water, the mixture was incubated at 65°C for 30 min. ForDNA extraction 750 �l of chloroform was added, mixed well, and centrifuged at14,000 rpm for 5 min in a Microfuge. The resultant upper phase was transferredto a clean tube with 420 �l of isopropanol and mixed gently. The tubes were thencooled on ice and spun in a Microfuge for 30 min at 14,000 rpm and 4°C.Following removal of the supernatant, the DNA pellet was washed with 75%ethanol and air dried. The DNA was then resuspended in RNase- and DNase-free water, quantified, diluted to 50 to 500 �g/ml, and used for PCR.
PCR amplification primers and conditions. The target gene loci and theirprimer names, primer sequences, primer locations in the M. tuberculosis H37Rvgenome (accession no. NC_000962) (9), and amplification product sizes and theprograms used to amplify are listed in Table 1. Primer pairs suited to evaluate thechosen genetic elements were created by using the DNASTAR program (DNA-STAR, Inc., Madison, Wis.) and sequence database information (http//:www.ncbi.nlm.nih.gov and http//:www.sanger.ac.uk/Projects/M_tuberculosis). Each PCRmixture was prepared with 25 �l of PCR Master (Roche Diagnostics, Indianap-olis, Ind.), 20 �l of water, 2.5 �l of dimethyl sulfoxide, 1 �l of each primer at 20�M, and 0.5 �l of DNA (equaling 25 to 250 ng). The sole difference was with the(additional) Rv3879c primer pair, which required 5 �l of dimethyl sulfoxide forgood amplification (final volume, 52.5 �l). PCR amplifications were performedin a Gene Amp PCR system 9700 (PE Applied Biosystems, Foster City, Calif.),using either program 1 (with an initial denaturation step of 5 min at 94°Cfollowed by 25 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C, andending with a final elongation step for 10 min at 72°C), program 2 (program 1 butwith an annealing temperature of 65°C), or program 3 (program 1 but with anannealing temperature of 50°C). All of the main MtbC PCR typing panel am-plifications used program 1. When DNA samples were limited or of low con-centration, the number of PCR cycles was increased to 35. PCR products and a100-bp ladder (Gibco BRL, Grand Island, N.Y.) were visualized by agarose(1.5%) gel electrophoresis and ethidium bromide staining. Images were capturedwith the Nighthawk imaging system (PDI Inc., Huntington Station, N.Y.) andQuality One software package (PDI Inc.). For this work all of the described PCRresults from mycobacteria included a positive control for amplification of thetarget locus as well as a positive control for the test DNA. Furthermore, allnegative and unexpected positive PCR results, as given in Table 2, were repeatedand confirmed at least once again.
DNA sequencing and PCR product restriction enzyme digest analysis. Directsequencing of PCR fragments was performed by the Cornell University BioRe-source Center (Ithaca, N.Y.) (http://www.brc.cornell.edu), using a BigDye Ter-minator kit (PE Applied Biosystems), and the output was analyzed with an ABI3700 DNA sequencer. The PCR amplification primers were also used as se-quencing primers, and a minimal single overlap from two directions for each wasusually achieved. Additional sequencing primers for gyrB and the 16S rRNA longfragment are listed in Table 1. The DNASTAR program was used to comparethe derived sequence data with DNA sequence information taken from theGenBank database. Comparative analysis of polymorphisms in the gyrA codon 95(gyrA95; G3C at nucleotide 7585 [7585G3C]) (nucleotide numberings are rel-ative to the plus strand of the M. tuberculosis H37Rv chromosome, accession no.NC_000962 [9]) and katG codons 203 and 463 (katG203 and katG463;2155501G3A and 2154722C3A, respectively) (7, 12, 17, 39) allows the segre-gation of the MtbC subspecies into four distinct genotypic groups: 1a, 1b, 2, and3. M. tuberculosis strains partition into either group 1b, 2, or 3; M. africanum
1638 HUARD ET AL. J. CLIN. MICROBIOL.
TABLE 1. Primers used in this study
Primer type and target locus Primer name Nucleotide sequence Locationa Size (bp) Programb
Main MtbC PCR typingpanel primer pairs
16S rRNA 16SRNAF 5� ACG GTG GGT ACT AGG TGT GGG TTT C 3� 1472650–674 543 116SRNAR 5� TCT GCG ATT ACT AGC GAC TCC GAC TTC A 3� 1473192–165
Rv0577 Rv0577F 5� ATG CCC AAG AGA AGC GAA TAC AGG CAA 3� 671164–190 786 1Rv0577R 5� CTA TTG CTG CGG TGC GGG CTT CAA 3� 671949–926
IS1561� (Rv3349c) IS1561F 5� GCT GGG TGG GCC CTG GAA TAC GTG AAC TCT 3� 3733670–699 943 1IS1561R 5� AAC TGC TCA CCC TGG CCA CCA CCA TTG ACT 3� 3754583–612
Rv1510 (RD4) Rv1510F 5� GTG CGC TCC ACC CAA ATA GTT GC 3� 1701578–600 1,033 1Rv1510R 5� TGT CGA CCT GGG GCA CAA ATC AGT C 3� 1702610–586
Rv1970 (RD7) Rv1970F 5� GCG CAG CTG CCG GAT GTC AAC 3� 2214244–264 1,116 1Rv1970R 5� CGC CGG CAG CCT CAC GAA ATG 3� 2215359–339
Rv3877/8 (RD1) Rv3877/8F 5� CGA CGG GTC TGA CGG CCA AAC TCA TC 3� 4356295–320 999 1Rv3877/8R 5� CTT GCT CGG TGG CCG GTT TTT CAG C 3� 4357269–293
Rv3120 (RD12) Rv3120F 5� GTC GGC GAT AGA CCA TGA GTC CGT CTC CAT 3� 3485555–584 404 1Rv3120R 5� GCG AAA AGT GGG CGG ATG CCA GAA TAG T 3� 3485958–931
Additional PCR primer pairsrpoB rpoBF 5� TCA AGG AGA AGC GCT ACG A 3� 760688–706 359 1
rpoBR 5� GGA TGT TGA TCA GGG TCT GC 3� 761047–028IS1081 1081F 5� TCG CGT GAT CCT TCG AAA CG 3� 1169369–388c 238 1
1081R 5� GCC GTT GCG CTG ATT GGA CC 3� 1169369–587c
MPB70 (Rv2875) MB1 5� GGC GAT CTG GTG GGC CCG 3� 3187117–134 489 2MB2 5� CGC CGG AGG CAT TAG CAC GCT 3� 3187605–585
IS1561�-2 2 IS1561F 5� GAC CTG ACG CCG CTG ACA C 3� 3754127–145 530 12 IS1561R 5� CAC CTA CAC CGC TTC CTG CC 3� 3754638–657
Rv3879c (RD1) Rv3879cF 5� TCT CCG GAA TGT CAT CTG GCT CCA GCA CAA 3� 4357958–987 1,000d 1Rv3879cR 5� GCA ACC CCG GCC ACG CCC GTT ACC 3� 4358957–934
Rv1510-2 (RD4) Y277-32F 5� GAC ATG TAC GAG AGA CGG CAT GAG 3� 1701290–313 1,031 1Y277-32R 5� AAT CCA ACA CGC AGC AAC CAG 3� 1702320–300
RD8 (5� junction) RD8-5�jnF 5� ATG CGC CAA CCG CCG TGT AGG 3� 4056590–610 956 1RD8-5�jnR 5� TGC CGG CCA GGT CCA GTT CAA AT 3� 4057545–523
Rv2073c (RD9) Rv2073cF 5� TCG CCG CTG CCA GAT GAG TC 3� 2330577–596 600 1Rv2073cR 5� TTT GGG AGC CGC CGG TGG TGA TGA 3� 2331171–148
Rv0222 (RD10) RD10iF 5� AGG GAT TCG GCG TTC GGG ATT CCT G 3� 265886–910 666 2RD10iR 5� GAT CGC GAT CGC CAA CGA TTC AGT GTA TG 3� 266551–523
Rv1257c (RD13) Rv1257cF 5� GGT GGC GAG CTG GAA TTC GTG AGA CAT TAC 3� 1404563–592 456 2Rv1257cR 5� CAT CGC CGA GGA GCG GAA TCT GAT GAT 3� 1405018–4992
Primers for gene sequencingand/or PCR-RFLP
oxyR285 oxyRF 5� CTA TGC GAT CAG GCG TAC TTG 3� 2725558–578 556 3oxyRR 5� GGT GAT ATA TCA CAC CAT A 3� 2726113–095
pncA57 pncAF 5� CAG GAG CTG CAA ACC AAC TCG 3� 2288680–700 664 1pncAR 5� GCT GGT CAT GTT CGC GAT CG 3� 2289343–324
hsp65631 hsp65F 5� ACC AAC GAT GGT GTG TCC AT 3� 528750–769 441 1hsp65R 5� CTT GTC GAA CCG CAT ACC CT 3� 529190–171
gyrB� gyrBF 5� TCG GAC GCG TAT GCG ATA TC 3� 5570–589 1,040 1gyrBR 5� ACA TAC AGT TCG GAC TTG CG 3� 6609–590
gyrA95 gyrAF 5� GGA GGT GCG CGA CGG GCT CAA G 3� 7427–448 354 2gyrAR 5� ACC CGG CCG TCG TAG TTA GGG ATG AAA TC 3� 7780–752
katG203 katG203F 5� GCC GGC GCC ATG GGT CTT ACC GAA AGT GTG 3� 2155273–302 370 2katG203R 5� CAA GAA GCT CTC ATG GGC GGA CCT GAT TGT 3� 2155642–613
katG463 katG463F 5� GAC GAG GTC GGC GAA GGA CAC TTT GA 3� 2154478–503 351 2katG463R 5� GGG CCG CTG GTC CCC AAG CAG AC 3� 2154828–806
16S rRNA (long) 16SLongF 5� TAA CAC ATG CAA GTC GAA CGG AAA GG 3� 1471895–920 1,436 116SLongR 5� ACT TCG TCC CAA TCG CCG ATC CCA CC 3� 1473331–305
Additional sequencingprimers
16S rRNA Seq 16S seqR 5� TTC ACG AAC AAC GCG ACA AAC CAC 3� 1472442–419gyrB Seq gyrB seqR 5� GCG GTT CGC TGA CCT TCA CCG AGA TCA C 3� 6335–307
a Coordinates as given by the M. tuberculosis H37Rv genome sequence, accession no. NC_000962 (reference 9).b See Materials and Methods.c Only one example of five IS1081 elements in the M. tuberculosis H37Rv genome is given.d Some M. tuberculosis strains amplified a 953-bp band.
VOL. 41, 2003 DIFFERENTIATION OF M. TUBERCULOSIS COMPLEX SUBSPECIES 1639
TA
BL
E2.
PCR
anal
ysis
ofm
ycob
acte
rial
gene
ticel
emen
ts
Stra
ina
Prev
ious
publ
icat
ion
(ref
eren
ce)
Mtb
Csu
bspe
cies
orM
OT
Tsp
ecie
sH
ost
Loc
atio
n
Res
ultb
for:
16S
rRN
Arp
oBR
v057
7IS
1081
MPB
70IS
1561
�R
v387
7/8
(RD
1)R
v151
0(R
D4)
Rv3
120
(RD
12)
Rv1
257c
(RD
13)
RV
1970
(RD
7)R
D8
Rv2
073c
(RD
9)R
v022
2(R
D10
)
217-
9413
“M.c
anet
tii”
Hum
anZ
uric
h�
��
��
��
��
c�
��
��
1772
7(1
16)
25“M
.can
ettii
”H
uman
Som
alia
��
��
��
��
�c
��
��
�96
-46
“M.c
anet
tii”
Hum
anF
ranc
e�
��
��
��
��
c�
��
��
97-4
88M
.tub
ercu
losi
sH
uman
The
Net
herl
ands
��
��
��
��
��
��
��
97-7
42M
.tub
ercu
losi
sH
uman
The
Net
herl
ands
��
��
��
��
��
��
��
97-8
03M
.tub
ercu
losi
sH
uman
The
Net
herl
ands
��
��
��
��
��
��
��
97-8
18M
.tub
ercu
losi
sH
uman
The
Net
herl
ands
��
��
��
��
��
��
��
97-1
177
M.t
uber
culo
sis
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�97
-128
9M
.tub
ercu
losi
sH
uman
The
Net
herl
ands
��
��
��
��
��
��
��
97-1
438
M.t
uber
culo
sis
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�W
M.t
uber
culo
sis
Hum
anU
nite
dSt
ates
��
��
��
��
��
��
��
CA
-43
(43)
25M
.tub
ercu
losi
sH
uman
Chi
na�
��
��
��
��
��
��
�C
A-6
5(6
5)25
M.t
uber
culo
sis
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�C
DC
1551
M.t
uber
culo
sis
Hum
anU
nite
dSt
ates
��
��
��
��
��
��
��
Cb3
.3M
.tub
ercu
losi
sH
uman
Uni
ted
Stat
es�
��
��
��
��
��
��
�20
01-1
225
M.t
uber
culo
sis
orM
.afr
ican
umsu
btyp
eII
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�
CA
-56
(56)
25M
.tub
ercu
losi
sH
uman
Cur
acao
��
��
��
��
��
��
��
97-6
6M
.tub
ercu
losi
sH
uman
The
Net
herl
ands
��
��
��
��
��
��
��
97-2
79M
.tub
ercu
losi
sH
uman
The
Net
herl
ands
��
��
��
��
��
��
��
97-1
503
M.t
uber
culo
sis
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�A
HT
N13
475
M.t
uber
culo
sis
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�M
tb21
M.t
uber
culo
sis
Hum
anU
nite
dSt
ates
��
��
��
��
��
��
��
Mtb
22M
.tub
ercu
losi
sH
uman
Uni
ted
Stat
es�
��
��
��
��
��
��
�97
0623
M.t
uber
culo
sis
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�A
TC
C27
294
M.t
uber
culo
sis
H37
Rv
Uni
ted
Stat
es�
��
��
��
��
��
��
�A
TC
C25
177
M.t
uber
culo
sis
H37
Ra
Uni
ted
Stat
es�
��
��
��
��
��
��
�A
TC
C51
910
M.t
uber
culo
sis
Hum
anU
nite
dSt
ates
��
��
��
��
��
��
��
NY
iso
M.t
uber
culo
sis
Hum
anU
nite
dSt
ates
��
��
��
��
��
��
��
CA
-105
(105
)25
M.t
uber
culo
sis
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�
1387
6(1
00)
25M
.afr
ican
umsu
btyp
eII
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�15
082
(92)
25M
.afr
ican
umsu
btyp
eII
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�15
199
(47)
25M
.afr
ican
umsu
btyp
eII
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�13
3/92
17M
.afr
ican
umsu
btyp
eII
Hum
anU
gand
a�
��
��
��
��
��
��
�17
8/92
17M
.afr
ican
umsu
btyp
eII
Hum
anU
gand
a�
��
��
��
��
��
��
�29
0/92
17M
.afr
ican
umsu
btyp
eII
Hum
anU
gand
a�
��
��
��
��
��
��
�11
67/9
317
M.a
fric
anum
subt
ype
IIH
uman
Uga
nda
��
��
��
��
��
��
��
AT
CC
2542
012
M.a
fric
anum
subt
ype
IH
uman
Sene
gal
��
��
��
��
��
��
��
AT
CC
3571
112
M.a
fric
anum
subt
ype
IH
uman
ND
d�
��
��
��
��
��
��
�12
55/9
317
M.a
fric
anum
subt
ype
IH
uman
Sier
raL
eone
��
��
��
��
��
��
��
1457
/93
17M
.afr
ican
umsu
btyp
eI
Hum
anSi
erra
Leo
ne�
��
��
��
��
��
��
�15
65/9
317
M.a
fric
anum
subt
ype
IH
uman
Sier
raL
eone
��
��
��
��
��
��
��
1567
/93
17M
.afr
ican
umsu
btyp
eI
Hum
anSi
erra
Leo
ne�
��
��
��
��
��
��
�17
902
(85)
25M
.afr
ican
umsu
btyp
eI
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�17
316
(6)
25M
.afr
ican
umsu
btyp
eI
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�
AT
CC
1942
244
M.m
icro
tiV
ole
Uni
ted
Kin
gdom
��
��
��
��
��
��
��
AT
CC
3578
2M
.mic
roti
Vol
eN
D�
��
��
��
��
��
��
�A
TC
C11
152
44M
.mic
roti
Vol
eU
nite
dK
ingd
om�
��
��
��
��
��
��
�97
-227
244
M.m
icro
tiH
uman
The
Net
herl
ands
��
��
��
��
��
��
��
1640 HUARD ET AL. J. CLIN. MICROBIOL.
97-2
257
44M
.mic
roti
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�15
496
44M
.mic
roti
Vol
eU
nite
dK
ingd
om�
��
��
��
��
��
��
�15
498
44M
.mic
roti
Vol
eU
nite
dK
ingd
om�
��
��
��
��
��
��
�16
420
44M
.mic
roti
Vol
eU
nite
dK
ingd
om�
��
��
��
��
��
��
�15
912
44M
.mic
roti
Lla
ma
Bel
gium
��
��
��
��
��
��
��
97-1
297
44M
.mic
roti
Fer
etT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�
Cip
1057
762
M.c
apro
eG
oat
Spai
n�
��
��
��
��
��
��
�
AT
CC
1921
0M
.bov
isC
owU
nite
dSt
ates
��
��
��
��
��
��
��
TN
5022
M.b
ovis
Hum
anU
nite
dSt
ates
��
��
��
��
��
��
��
AT
CC
3572
5M
.bov
isN
DN
D�
��
��
��
��
��
��
�A
TC
C35
726
M.b
ovis
Cow
ND
��
��
��
��
��
��
��
AT
CC
3573
0M
.bov
isC
owN
D�
��
��
��
��
��
��
�20
01-8
31M
.bov
isH
uman
The
Net
herl
ands
��
��
��
��
��
��
��
CA
-73
(73)
25M
.bov
isC
owT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�C
A-1
17(1
17)
25M
.bov
isC
owA
rgen
tina
��
��
��
��
��
��
��
CA
-126
(126
)25
M.b
ovis
Cow
Arg
entin
a�
��
��
��
��
��
��
�C
A-1
30(1
30)
25M
.bov
isC
owT
heN
ethe
rlan
ds�
��
��
��
��
��
��
�
BC
Gt
M.b
ovis
BC
GV
acci
nePa
steu
r-T
unis
��
��
��
��
��
��
��
r4-9
3M
.bov
isB
CG
ND
ND
��
��
��
��
��
��
��
AT
CC
2729
0M
.bov
isB
CG
Vac
cine
Cop
enha
gen
��
��
��
��
��
��
��
AT
CC
3573
6M
.bov
isB
CG
Vac
cine
Bra
zil
��
��
��
��
��
��
��
AT
CC
3573
7M
.bov
isB
CG
Vac
cine
Japa
n�
��
��
��
��
��
��
�T
N10
130
M.b
ovis
BC
GH
uman
Uni
ted
Stat
es�
��
��
��
��
��
��
�
NY
S99
76M
.abs
cess
usR
ef.e
Uni
ted
Stat
es�
��
��
��
��
��
H15
882
M.a
bsce
ssus
Hum
anU
nite
dSt
ates
��
��
��
��
��
�H
2147
9M
.abs
cess
usH
uman
Uni
ted
Stat
es�
��
��
��
��
��
T61
715
M.a
bsce
ssus
Hum
anU
nite
dSt
ates
��
��
��
��
��
�
MA
C(B
J)M
.avi
umco
mpl
exH
uman
Uni
ted
Stat
es�
��
��
��
��
��
MA
C(F
L)
M.a
vium
com
plex
Hum
anU
nite
dSt
ates
��
��
��
��
��
�A
TC
C19
074
M.a
vium
subs
p.av
ium
Hum
anU
nite
dSt
ates
��
��
��
��
��
�A
TC
C25
291
M.a
vium
subs
p.av
ium
Chi
cken
ND
��
��
��
��
��
�A
TC
C35
781
M.a
vium
subs
p.av
ium
ND
ND
��
��
��
��
��
�98
0084
7M
.avi
umsu
bsp.
aviu
mG
oose
Bel
gium
��
��
��
��
��
�96
0113
8M
.avi
umsu
bsp.
aviu
mH
uman
The
Net
herl
ands
��
��
��
��
��
�A
TC
C19
077
M.a
vium
subs
p.in
trac
ellu
lare
Hum
anN
D�
��
��
��
��
��
AT
CC
1395
0M
.avi
umsu
bsp.
intr
acel
lula
reN
DN
D�
��
��
��
��
��
9601
103
M.a
vium
subs
p.in
trac
ellu
lare
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
EG
M.c
helo
nae
Hum
anU
nite
dSt
ates
��
��
��
��
��
�PB
M.c
helo
nae
Hum
anU
nite
dSt
ates
��
��
��
��
��
�N
YS
IS01
36M
.for
tuitu
mR
ef.
Uni
ted
Stat
es�
��
��
��
��
��
W37
396
M.f
ortu
itum
Hum
anU
nite
dSt
ates
��
��
��
��
��
�H
4958
1M
.for
tuitu
mH
uman
Uni
ted
Stat
es�
��
��
��
��
��
NY
SIS
9775
M.f
ortu
itum
Ref
.U
nite
dSt
ates
��
��
��
��
��
�M
LM
.for
tuitu
mH
uman
Uni
ted
Stat
es�
��
��
��
��
��
YG
M.f
ortu
itum
Hum
anU
nite
dSt
ates
��
��
��
��
��
�B
DM
.gor
dona
eH
uman
Uni
ted
Stat
es�
��
��
��
��
��
GM
M.g
ordo
nae
Hum
anU
nite
dSt
ates
��
��
��
��
��
�
Con
tinue
don
follo
win
gpa
ge
VOL. 41, 2003 DIFFERENTIATION OF M. TUBERCULOSIS COMPLEX SUBSPECIES 1641
TA
BL
E2—
Con
tinue
d
Stra
ina
Prev
ious
publ
icat
ion
(ref
eren
ce)
Mtb
Csu
bspe
cies
orM
OT
Tsp
ecie
sH
ost
Loc
atio
n
Res
ultb
for:
16S
rRN
Arp
oBR
v057
7IS
1081
MPB
70IS
1561
�R
v387
7/8
(RD
1)R
v151
0(R
D4)
Rv3
120
(RD
12)
Rv1
257c
(RD
13)
RV
1970
(RD
7)R
D8
Rv2
073c
(RD
9)
Rv0
222
(RD
10)
-
9004
19M
.kan
sasi
iH
uman
The
Net
herl
ands
��
��
��
��
��
�20
00-1
049
M.k
ansa
sii
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
2000
-145
4M
.kan
sasi
iT
apw
ater
Bel
gium
��
��
��
��
��
�20
00-1
457
M.k
ansa
sii
Bio
film
Ger
man
y�
��
��
��
��
��
2000
-145
8M
.kan
sasi
iT
apw
ater
Ger
man
y�
��
��
��
��
��
2000
-145
9M
.kan
sasi
iT
apw
ater
Bel
gium
��
��
��
��
��
�20
00-1
461
M.k
ansa
sii
Toi
let
Bel
gium
��
��
��
��
��
�20
00-1
471
M.k
ansa
sii
Env
iron
men
tIt
aly
��
��
��
��
��
�20
00-1
493
M.k
ansa
sii
Wat
erG
erm
any
��
��
��
��
��
�20
00-1
495
M.k
ansa
sii
Hot
wat
erC
zech
Rep
ublic
��
��
��
��
��
�
J186
98M
.mal
moe
nse
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
AT
CC
927
M.m
arin
umF
ish
Uni
ted
Stat
es�
��
��
��
��
��
9801
810
M.m
arin
umH
uman
The
Net
herl
ands
��
��
��
��
��
�99
0036
M.m
arin
umH
uman
The
Net
herl
ands
��
��
��
��
��
�
xsim
M.s
imia
eN
DU
nite
dSt
ates
��
��
��
��
��
�
AT
CC
2303
8M
.sm
egm
atis
ND
ND
��
��
��
��
��
�m
c2M
.sm
egm
atis
mc2
155
Uni
ted
Stat
es�
��
��
��
��
��
SCC
74/3
1M
.szu
lgai
Hum
anT
heN
ethe
rlan
ds�
��
��
��
��
��
myc
941
M.x
enop
iH
uman
The
Net
herl
ands
��
��
��
��
��
�SR
-BL
M.x
enop
iH
uman
Uni
ted
Stat
es�
��
��
��
��
��
aU
nder
lined
stra
ins
wer
ere
clas
sifie
din
this
stud
yan
dw
ere
orig
inal
lyde
sign
ated
M.a
fric
anum
[no
subt
ype]
(138
76an
d15
082)
,M.b
ovis
(200
1-12
25,1
5199
,179
02,1
7316
,and
TN
1013
0),o
rM
.mic
roti
(AT
CC
3578
1an
dxs
im).
Iden
tifica
tion
num
bers
from
refe
renc
e25
are
inbo
ldfa
ce.
b�
,neg
ativ
efo
ra
stro
ngba
ndof
the
corr
ect
size
;�,p
ositi
vefo
rsu
cha
band
.c
RD
can
part
ially
over
laps
with
RD
12(s
eere
fere
nce
6).
dN
D,n
oda
ta.
eR
ef.,
refe
renc
est
rain
.
1642 HUARD ET AL. J. CLIN. MICROBIOL.
subtype II strains fall into either MtbC group 1b or 2; “M. canettii” strains areMtbC group 1b; and M. bovis, M. bovis BCG, M. caprae, M. microti, and M.africanum subtype I strains fall into group 1a. For the strains indicated in Table3, the determination of MtbC grouping was done by sequence analysis of thegyrA95 PCR fragment and PCR-RFLP of the katG203 and katG463 amplicons withthe restriction enzyme BstNI (New England Biolabs, Beverly, Mass.). BstNIdigestion of the katG203 PCR product amplified from MtbC group 1b, 2, and 3isolates produces two DNA fragments (230 and 140 bp), while that from MtbCgroup 1a strains remains uncut. Similarly, BstNI digestion of the katG463 PCRproduct produces three DNA fragments (12, 61, and 278 bp) from MtbC group2 and 3 isolates, while that from MtbC group 1a and 1b strains is cut into fourDNA fragments (12, 61, 104, and 174 bp). Digest products were visualized byagarose (4%) gel electrophoresis with ethidium bromide staining. Nucleotidesubstitutions in pncA, oxyR�, hsp65, and gyrB have also been used to identifyspecific MtbC members. The pncA codon 57 SNP (pncA57; 2289071G3C) seg-regates M. bovis (strict sense) and M. bovis BCG from the other MtbC subspecies(36), while the oxyR� pseudogene bears a position 285 SNP (oxyR285;2725801C3T) that differentiates M. bovis (strict sense), M. bovis BCG, and M.caprae isolates from the other MtbC members (31, 38). As a result, the pncA57
nucleotide usage relative to that of oxyR285 singles out M. caprae strains. Simi-larly, a position 631 SNP in the hsp65 gene (hsp65631; 529004C3T) is restrictedto “M. canettii” strains, differentiating them from the other MtbC subspecies(13), while unique SNPs in a �1-kb section of the gyrB gene (gyrB� [� representsthe polymorphic amplicon]) allow M. africanum subtype I (gyrBAf, 6446G3T),M. microti (gyrBMic, 5617C3T and 6446G3T), M. bovis (strict sense) (gyrBBv,5752G3A, 6406C3T, and 6446G3T), and M. caprae (gyrBCp, 5752G3A,6307T3G, and 6446G3T), to be discriminated from each other and from M.tuberculosis or M. africanum subtype II (gyrBTb) (24, 30). PCR-RFLP of theoxyR285, hsp65631, and gyrB� PCR fragments was done to suggest subspeciesidentity, for the strains indicated in Table 3, as previously described (13, 31, 38).Likewise, PCR-RFLP of the pncA57 PCR product was done with BstEII (NewEngland Biolabs), a restriction enzyme that cuts the M. bovis (strict sense)pncA57 amplicon into two fragments (170 and 494 bp) and cuts pncA57 from mostother MtbC strains into three fragments (103, 170, and 391 bp) (Table 3).Additional digestion of the gyrB� PCR fragment (to probe for the 6406C3TSNP) with the MaeIII restriction enzyme (Roche Diagnostics) was done to cutthe gyrBBv amplicon into three fragments (306, 331, and 403 bp) and to cut thegyrB� from the remaining MtbC into four DNA fragments (123, 208, 306, and403 bp) (Table 3). Direct sequencing of katG203, katG463, oxyR285, pncA57,hsp65631, and gyrB� PCR products was done to confirm the respective PCR-RFLP results from a sample of MtbC strains.
RESULTS
Various chromosomal loci were evaluated to ascertain theirusefulness as part of an MtbC PCR typing panel.
Selective amplification of the mycobacterial 16S rRNA gene.For the present work it was necessary to have a means ofconfirming the presence of mycobacterial DNA. Previously,species-specific nucleotide polymorphisms in the 16S rRNAgene have been used to determine Mycobacterium species iden-tity (37, 41). For pan-genus PCR amplification of the myco-bacterial 16S rRNA, a primer pair was designed to consensusnonpolymorphic segments of the 16S rRNA GenBank genesequences from two MtbC subspecies and several MOTT spe-cies. These primers were successful in amplifying a DNA frag-ment from all mycobacterial isolates (n � 115) in the collection(Table 2). Likewise, an alternative PCR primer pair to therpoB gene, designed in the same manner as the 16S rRNAprimers, amplified DNA from all tested mycobacteria (n �115) (Table 2). Importantly, the evaluated MOTT species in-cluded the nontuberculous organisms most commonly encoun-tered in clinical practice (19). To further validate species iden-tity, the PCR amplification products from several MtbC strains(n � 21) and all MOTT strains (n � 44) were sequenced andconfirmed to have the 16S rRNA nucleotide sequence reportedfor that species in GenBank (data not shown). Amplification
for the 16S rRNA was chosen to provide the positive controlwhen evaluating mycobacteria by PCR.
The Rv0577 gene is restricted to the MtbC subspecies. On-going tuberculosis-related projects revealed the potential ofRv0577 as an MtbC-restricted gene. Primers were generated toamplify the full coding region of Rv0577 and were then used totest the collected mycobacteria (n � 115). Rv0577 was specif-ically and consistently amplified from all MtbC subspecies andstrains tested (n � 71), whereas none of the MOTT strains (n� 44) showed a PCR fragment (Table 2). To establish theappropriate amplification of Rv0577, the PCR product fromM. tuberculosis H37Ra was sequenced (data not shown). More-over, PCR amplification results for IS1081 and MPB70, geneticelements believed to be absent from most or all MOTT species(27), paralleled the MtbC-restricted pattern of Rv0577 (n �115 mycobacteria tested) (Table 2). These results support thatRv0577 is a genotypic marker for the MtbC and that it can beused to distinguish the MtbC subspecies from MOTT species.
Absence of the IS1561� element differentiates M. microtifrom the other MtbC subspecies. Previously, Gordon et al. (15)noted that PCR amplification for the transposase pseudogenefragment IS1561� (Rv3349c) was positive for all MtbC isolatestested except for their single M. microti (OV254) strain, andthey postulated that it might prove to be a M. microti-specificmarker. For this study, primers to the IS1561� element weregenerated and the collected MtbC strains were evaluated. TheIS1561� PCR fragment was found to be absent from all of theM. microti strains (n � 10) but was present in the other eval-uated MtbC isolates (n � 61) (Table 2). The identity of theIS1561� amplicon was verified by sequence analysis of the PCRfragment from M. tuberculosis H37Ra. In addition, amplifica-tion with a previously described IS1561� primer pair (15) con-firmed the lack of IS1561� amplicons from the M. microtistrains (n � 71 MtbC strains tested) (data not shown). The M.microti strains in this collection were isolated from diversesources (including llama, ferret, and vole, as well as human)and locations (43). Moreover, a very recent paper describingnew M. microti LSPs also found the IS1561� locus to be deleted(5). Therefore, these data support that the absence of IS1561�is a good genotypic indicator for M. microti.
Absence of the Rv1510 gene differentiates M. bovis and M.bovis BCG from the other MtbC subspecies. Previous studiesindicated that PCR primers generated to the RD4 locus coulddistinguish M. bovis (strict sense) and M. bovis BCG from therest of the MtbC subspecies (4, 5, 14). In order to confirm M.bovis subspecies identity, primers were generated to Rv1510, agene located internal to the RD4 deletion. Amplification ex-periments confirmed that Rv1510 was absent in all of theevaluated M. bovis strains (n � 10) and M. bovis BCG strains(n � 6) but was present in the other MtbC isolates of thecollection (n � 55) (Table 2). The identity of the Rv1510amplification product generated from M. tuberculosis H37Rawas proven by sequence analysis. Furthermore, using alterna-tive Rv1510 primers described previously (14), M. bovis and M.bovis BCG were also segregated from the remaining MtbCstrains (n � 71 MtbC isolates tested) (data not shown). There-fore, these results support that RD4 is a genotypic marker forM. bovis (strict sense) and M. bovis BCG and that the absenceof Rv1510 can be used to differentiate them from the otherMtbC subspecies.
VOL. 41, 2003 DIFFERENTIATION OF M. TUBERCULOSIS COMPLEX SUBSPECIES 1643
TABLE 3. Single-nucleotide polymorphism analysis of MtbC isolatesa
Strainb MtbC subspecies MtbCgrouping
katG203
(2155501)katG463
(2154722)gyrA95
(7585)
gyrB�hsp65631
(529004)oxyR285
(2725801)pncA57
(2289071)5671 5752 6307 6406 6446
217-94 “M. canettii” 1b G A C C G T C G T C G17727 “M. canettii” 1b G A C C G T C G T C G96-46 “M. canettii” 1b G A C C G T C G T C G
97-488 M. tuberculosis 1b G A C C G T C G C C G97-742 M. tuberculosis 1b G A C C G T C G C C G97-803 M. tuberculosis 1b G A C C G T C G C C G97-818 M. tuberculosis 1b G A C C G T C G C C G97-1177 M. tuberculosis 1b G A C C G T C G C C G97-1289 M. tuberculosis 1b G A C C G T C G C C G97-1438 M. tuberculosis 1b G A C C G T C G C C GW M. tuberculosis 1b G A C C G T C G C C GCA-43 M. tuberculosis 1b G A C C G T C G C C GCA-65 M. tuberculosis 1b G A C C G T C G C C GCDC1551 M. tuberculosis 2 G C C C G T C G C C GCb3.3 M. tuberculosis 2 G C C C G T C G C C G2001-1225 M. tuberculosis or M.
africanum subtype II2 G C C C G T C G C C G
97-56 M. tuberculosis 2 G C C C G T C G C C G97-66 M. tuberculosis 2 G C C C G T C G C C G97-279 M. tuberculosis 2 G C C C G T C G C C G97-1503 M. tuberculosis 2 G C C C G T C G C C GAH TN13475 M. tuberculosis 2 G C C C G T C G C C GMtb21 M. tuberculosis 2 G C C C G T C G C C GMtb22 M. tuberculosis 2 G C C C G T C G C C G970623 M. tuberculosis 2 G C C C G T C G C C GH37Rv M. tuberculosis 3 G C G C G T C G C C GH37Ra M. tuberculosis 3 G C G C G T C G C C GATCC 51910 M. tuberculosis 3 G C G C G T C G C C GNY iso M. tuberculosis 3 G C G C G T C G C C GCA-105 M. tuberculosis 3 G C G C G T C G C C G
13876 M. africanum subtype II 1b G A C C G T C G C C G15082 M. africanum subtype II 1b G A C C G T C T C C G15199 M. africanum subtype II 1b G A C C G T C G C C G133/92 M. africanum subtype II 2 G C C C G T C G C C G178/92 M. africanum subtype II 2 G C C C G T C G C C G290/92 M. africanum subtype II 2 G C C C G T C G C C G1167/93 M. africanum subtype II 2 G C C C G T C G C C G
ATCC 25420 M. africanum subtype I 1a A A C C G T C T C C GATCC 35711 M. africanum subtype I 1a A A C C G T C T C C G1255/93 M. africanum subtype I 1a A A C C G T C T C C G1457/93 M. africanum subtype I 1a A A C C G T C T C C G1565/93 M. africanum subtype I 1a A A C C G T C T C C G1567/93 M. africanum subtype I 1a A A C C G T C T C C G17902 M. africanum subtype I 1a A A C C G T C T C C G17316 M. africanum subtype I 1a A A C C G T C T C C G
ATCC 19422 M. microti 1a A A C T G T C T C C GATCC 35782 M. microti 1a A A C T G T C T C C GATCC 11152 M. microti 1a A A C T G T C T C C G9402272 M. microti 1a A A C T G T C T C C G9702257 M. microti 1a A A C T G T C T C C G15496 M. microti 1a A A C T G T C T C C G15498 M. microti 1a A A C T G T C T C C G16420 M. microti 1a A A C T G T C T C C G15912 M. microti 1a A A C T G T C T C C G97-1297 M. microti 1a A A C T G T C T C C G
Cip 105776 M. caprae 1a A A C C A G C T C T G
ATCC 19210 M. bovis 1a A A C C A T T T C T CTN5022 M. bovis 1a A A C C A T T T C T C
Continued on following page
1644 HUARD ET AL. J. CLIN. MICROBIOL.
The Rv1970 gene differentiates M. tuberculosis, M. africanumsubtype II, and “M. canettii” from the other MtbC subspecies.Rv1970 (lprM) is a gene located internal to RD7 (4, 14). Inprior studies, PCR analysis revealed RD7 to be present in all ofthe tested M. tuberculosis and “M. canettii” strains, as well assome M. africanum isolates, but not in the evaluated M. mi-croti, M. bovis, M. bovis BCG, and M. caprae strains (7, 14, 29).Primers were generated to amplify Rv1970, and the PCR re-sults were positive for M. tuberculosis (n � 26), M. africanumsubtype II (n � 7), and “M. canettii” (n � 3) strains, while thegene from MtbC group 1a subspecies (n � 35) failed to amplify(Table 2). To ensure that Rv1970 was correctly amplified, thePCR product generated from M. tuberculosis H37Ra was se-quenced and shown to be Rv1970 (data not shown). In addi-tion, the RD7 LSP is absent concomitantly with the RD8 andRD10 locus deletions and is often, but not in all cases, associ-ated with the loss of the RD9 locus (7, 29). Primer pairs thatamplify either the RD9-internal gene Rv2073c or within theRD10 deletion Rv0222 gene or the RD8 deletion intergenicregion of Rv3616c and Rv3617 each segregated the evaluatedMtbC isolates identically to the Rv1970 PCR (n � 71 MtbCstrains evaluated) (Table 2). Therefore, amplification for theseloci supported the determined Rv1970 MtbC subspecies seg-regation, and amplification for Rv1970 was chosen for inclu-sion in the MtbC PCR typing panel.
Absence of the Rv3877 and Rv3878 genes of the RD1 locusdifferentiates M. bovis BCG from the other MtbC subspecies.The RD1 locus was found previously to be selectively absent inall M. bovis BCG isolates (4, 14), and the presence of RD1 hasbeen shown to be a good negative marker for M. bovis BCG(35, 40). For this study, a PCR primer pair was designed toamplify within the RD1 deletion genes Rv3877 and Rv3878(Rv3877/8). PCR testing of these primers revealed that, withthe exception of the M. bovis BCG isolates (n � 6), all of thetested MtbC strains (n � 65) amplified a specific DNA frag-ment (Table 2). Sequence analysis of this PCR product gener-ated from M. tuberculosis H37Ra confirmed it as Rv3877/8
(data not shown). As a result, this element appeared to providea good target for M. bovis BCG identification. A second RD1primer pair that amplifies a portion of Rv3879c was also eval-uated. These primers failed to amplify from all M. bovis BCGisolates, as expected, but also failed to amplify from M. tuber-culosis strain CA-56, an isolate that has been described to havean undefined LSP that partially overlaps RD1 (reference 7 anddata not shown). In addition, the M. tuberculosis strains 97-803,97-117, and 97-1438 were each found to have a novel andidentical 57-bp deletion within Rv3879c (nucleotides 4368661to 4368707 relative to the M. tuberculosis H37Rv genome se-quence [9]) that truncates the predicted protein by 19 aminoacids while maintaining the codon frame (data not shown).
The Rv3120 gene differentiates M. bovis, M. bovis BCG,M. caprae, and “M. canettii” from the other MtbC members.RDcan is a unique “M. canettii”-specific deletion that partiallyoverlaps with RD12, both of which include the gene Rv3120(7). To further segregate members of the MtbC, PCR primerswere generated to amplify within Rv3120. A PCR fragment ofthe correct size was detected from the evaluated M. tubercu-losis (n � 26), M. africanum subtype I (n � 8), M. africanumsubtype II (n � 7), and M. microti (n � 10) strains but not fromthe M. bovis (n � 10), M. bovis BCG (n � 6), M. caprae (n �1), and “M. canettii” (n � 3) strains (Table 2). Sequence anal-ysis of the Rv3120 PCR product from M. tuberculosis H37Rafurther confirmed it as Rv3120 (data not shown). In the pro-cess of building the MtbC PCR typing panel, a primer pair tothe RD13 deletion gene Rv1257c was also tested. The Rv1257cprimers exhibited the same MtbC amplification profile asRv3120 but with the distinct ability to amplify for “M. canettii”(n � 71 MtbC isolates tested) (Table 2), thereby supportingthe selectivity of the Rv3120 amplification results.
The composite PCR panel provides a unique pattern to typeindividual MtbC subspecies. The optimized and thoroughlyevaluated primers to the loci 16S rRNA, Rv0577, IS1561�,Rv1510, Rv1970, Rv3877/8, and Rv3120 (chosen for their re-spective amplification profiles from the evaluated mycobacte-
TABLE 3—Continued
Strainb MtbC subspecies MtbCgrouping
katG203
(2155501)katG463
(2154722)gyrA95
(7585)
gyrB�hsp65631
(529004)oxyR285
(2725801)pncA57
(2289071)(5671) (5752) (6307) (6406) (6446)
ATCC 35725 M. bovis 1a A A C C A T T T C T CATCC 35726 M. bovis 1a A A C C A T T T C T CATCC 35730 M. bovis 1a A A C C A T T T C T C2001-831 M. bovis 1a A A C C A T T T C T CCA-73 M. bovis 1a A A C C A T T T C T CCA-117 M. bovis 1a A A C C A T T T C T CCA-126 M. bovis 1a A A C C A T T T C T CCA-130 M. bovis 1a A A C C A T T T C T C
BCGt M. bovis BCG 1a A A C C A T T T C T Cr4-93 M. bovis BCG 1a A A C C A T T T C T CATCC 27290 M. bovis BCG 1a A A C C A T T T C T CATCC 35736 M. bovis BCG 1a A A C C A T T T C T CATCC 35737 M. bovis BCG 1a A A C C A T T T C T CTN10130 M. bovis BCG 1a A A C C A T T T C T C
a Coordinates as given by the M. tuberculosis H37Rv genome sequence, accession no. NC_000962 (see reference 9).b Underlined strains were reclassified in this study and originally designated M. africanum [no subtype] (13876 and 15082), or M. bovis (2001-1225, 15199, 17902,
17316, and TN10130).
VOL. 41, 2003 DIFFERENTIATION OF M. TUBERCULOSIS COMPLEX SUBSPECIES 1645
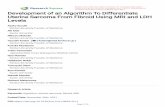
ria) form the MtbC PCR typing panel (Table 1). Figure 1illustrates the representative PCR amplification pattern foreach MtbC grouping and a single MOTT (M. avium subsp.avium) with this set of seven primer pairs. With the exceptionof M. tuberculosis and M. africanum subtype II (which wereidentical), this panel gave a unique pattern of DNA fragmentsfor each MtbC subspecies. When the MtbC PCR typing panelwas applied to the DNAs from the collected MOTT strains (n� 44) (Table 2), only the 16S rRNA primer pairs amplified,thereby giving the MOTT samples a single pattern. Table 4summarizes the distinct patterns whereby the amplificationresult (positive or negative) for each MtbC subspecies and thetested MOTT species is indicated. Based upon the presence ofthe expected PCR fragment, for each numerically assignedlocus, an algorithm is proposed for a MOTT and each MtbCsubspecies. As examples, a MOTT which amplifies only for the16S rRNA in lane 1 has a profile of 1, while M. tuberculosis andM. africanum subtype II, which amplify for all loci in the MtbCPCR typing panel, have a profile of 1234567.
MtbC strains which did not give the expected MtbC PCR
typing panel pattern. Throughout the course of this study, afew mycobacterial isolates as designated conflicted with theexpected results with the MtbC PCR typing panel as well asone or more of the following: PCR-RFLP analysis, 16S rRNAsequencing, amplification using the corroborative primer pairs,and/or published spoligotype data (25, 31, 45). Each of theseisolates was reclassified in the final synopsis and indicated byan underlined name (with reference to the prior designation)in Tables 2 and 3. For instance, a single “M. microti” isolate(ATCC 35781) in the collection had the MtbC PCR typingpanel profile of 1, the MOTT profile. This isolate failed toamplify for the additional MtbC-specific IS1081 and MPB70elements, and sequence determination of a 1,436-bp fragmentof the 16S rRNA from this isolate found it to match perfectly tothat of M. avium subsp. avium (accession no. AF306455). This“M. microti” strain was purchased directly at a time that no M.avium subsp. avium strains were present in the testing labora-tory and so was reclassified as M. avium subsp. avium (strainATCC 35781 in Table 2). As further examples, two strainsdetermined phenotypically and given as “M. bovis” human
FIG. 1. The composite MtbC PCR typing panel. Illustrated are the typical MtbC PCR panel typing results for a single representative of eachMtbC subspecies as well as MOTT (M. avium subsp. avium is shown). A total of 71 MtbC isolates and 44 MOTT isolates were tested. PCR productsand a 100-bp ladder were visualized by agarose gel electrophoresis and ethidium bromide staining. Images were captured with the Nighthawkimaging system (PDI Inc.) and Quality One software package (PDI Inc.). Lanes: 1, 16S rRNA; 2, Rv0577; 3, IS1561�; 4, Rv1510; 5, Rv1970; 6,Rv3877/8; 7, Rv3120. Unlabeled lanes in each panel contain the 100-bp ladder.
1646 HUARD ET AL. J. CLIN. MICROBIOL.
isolates did not yield the expected M. bovis profile 1236 (strains15199 and 2001-1225 in Tables 2 and 3) when tested by thePCR panel but gave the profile 1234567 and were determinedto be either M. africanum subtype II or M. tuberculosis. Supportfor reassignment came from their wild-type nucleotide usage inoxyR285 and pncA57 and for having gyrBTb. Furthermore, se-quence determination of katG203, katG463, and gyrA95 revealedstrain 15199 to be MtbC group 1b, while strain 2001-1225 wasMtbC group 2. Both groups are known to include M. tubercu-losis and M. africanum subtype II strains but not M. bovis (12,17, 39). The spoligotype of strain 15199 (25 [shown as strain47]) corresponded to the M. africanum subtype II spoligotypepatterns illustrated by Niemann et al. (31). The spoligotype ofstrain 2001-1225 was unavailable. Therefore, for this studystrain 15199 was designated M. africanum subtype II, whilestrain 2001-1225 could not be definitively identified and so isgiven as M. tuberculosis or M. africanum subtype II. The MtbCPCR typing panel also identified a probable cross-contamina-tion and confirmed its correction. Here, one M. bovis BCGstrain (BCGt) maintained in culture gave the expected 123profile from an earlier DNA sample, but a later DNA prepa-ration gave the 1234567 profile of M. tuberculosis or M. africa-num subtype II. Restarting the culture from stored stocks re-capitulated the 123 profile of M. bovis BCG (data not shown).These examples illustrate the usefulness of the MtbC PCRtyping panel for intralaboratory control practices and the man-agement of MtbC cultures. Importantly, there was near-com-plete concordance between the MtbC PCR panel-determinedMtbC subspecies identities (Table 2) and the known MtbCsubspecies-defining SNPs (Table 3). To our knowledge, thisstudy provides the most comprehensive comparative analysisand validation of the known MtbC-defining SNPs. However,M. africanum subtype II strain 15082 (25 [shown as strain 92]),which is otherwise typical by all other criteria (Tables 2 and 3),was not gyrBTb but rather was gyrBAf by both PCR-RFLP andsequence analysis and is a noteworthy exception.
DISCUSSION
This work describes the development of a simple, straight-forward, rapid, and specific PCR-based typing method thatunambiguously differentiates individual subspecies of theMtbC and segregates them from various clinically importantMOTT species. This MtbC PCR typing panel evaluates inde-
pendent loci that vary due to unidirectional chromosomal re-gion deletions, and, as a result, our data were in close concor-dance with recent independently derived RD data (5, 7, 29). Assuch, this protocol is an improvement over current phenotypicand molecular-based MtbC subspecies-determining strategies.However, an inherent concern of RD locus PCR targetingstrategies for MtbC subspecies determination is the potentialfor independent strain-specific deletions that overlap othersubspecies-specific or lineage-specific LSPs. Emerging RD locimay result in uncommon isolates with novel strain-specificMtbC PCR typing panel profiles or rare outliers that give theMtbC PCR typing panel profile of another MtbC subspecies.In both cases, alternative PCR target amplifications (such asthose provided in Table 1), SNP-based subspecies determina-tion, spoligotyping, or phenotypic characterization may be war-ranted. Alternatively, to compensate for such variants or novelMtbC subspecies, the MtbC PCR typing panel may be ex-panded to incorporate new target loci so as to increase itsdiscriminatory power.
The major limitation of our MtbC PCR typing panel is thatit cannot differentiate M. tuberculosis from M. africanum sub-type II. Compared to M. tuberculosis, M. africanum subtype IIis known to have certain unique phenotypic and biogeograph-ical features (17, 22, 30, 32), and yet we were unable to identifyany chromosomal differences to set these two apart. In fact, tothis point no single DNA-based protocol has been able topositively identify all M. africanum subtype II strains on itsown. Indeed, there is a growing consensus that M. africanumsubtype II strains are really simply clustered biovariants of M.tuberculosis with divergent properties in phenotypic analyses(7, 35). The facts that all M. africanum subtype II strains testedto date also lacked the “modern” M. tuberculosis deletionTbD1 (7) and that M. africanum subtype II isolates do not forma single lineage but can fall into either MtbC genotypic group1b or group 2 by katG203, katG463, and gyrA95 determinations(Table 3) support this view. If previously characterized M.africanum subtype II isolates actually represent atypical M.tuberculosis strains, then the MtbC PCR typing panel groupedthese isolates appropriately, reiterating the advantages of thistyping approach.
The MtbC PCR typing panel was originally intended for usein the confirmation of MtbC subspecies identity and as anintralaboratory control for cross-species contamination andother laboratory errors. For these uses the MtbC typing panel
TABLE 4. An algorithm to identify individual MtbC subspeciesa
Organism(s)
Result for the following locus (lane in Fig. 1):
Profile16S rRNA(1)
Rv0577(2)
IS1561�(3)
Rv1510(4)
Rv1970(5)
Rv3877/8(6)
Rv3120(7)
M. tuberculosis � � � � � � � 1234567M. africanum subtype II � � � � � � � 1234567“M. canettii” � � � � � � � 123456M. africanum subtype I � � � � � � � 123467M. caprae � � � � � � � 12346M. bovis � � � � � � � 1236M. bovis BCG � � � � � � � 123M. microti � � � � � � � 12467MOTT � � � � � � � 1
a Based on the results shown in Fig. 1.
VOL. 41, 2003 DIFFERENTIATION OF M. TUBERCULOSIS COMPLEX SUBSPECIES 1647
has been of great value, revealing several erroneously desig-nated MtbC strains. In the future, the MtbC PCR typing panelmay be applied to determine subspecies of stored clinical andagricultural isolates in retrospective epidemiological studiesthat may unveil previously unknown correlates of infection,at-risk populations, animal reservoirs, and nuances of clinicalcourse and treatment response, as well as pathogen geographicrange and spread over time. The MtbC PCR typing panel mayalso be useful for the typing of new clinical isolates, especiallyin areas where the geographic distributions of various MtbCsubspecies may overlap, as is potentially the case with M. tu-berculosis, M. africanum subtype II, M. africanum subtype I, M.bovis, and “M. canettii” in Africa. This simple and rapid typingprotocol could also help to determine the optimal therapeuticstrategy for individual cases in areas of high tuberculosis inci-dence where the diagnosis of tuberculosis is made more diffi-cult by human immunodeficiency virus coinfection and/orwhere the distinction from MOTT species is of the most im-portance. The MtbC PCR typing panel may also aid the diag-nosis of M. bovis BCG dissemination as a complication of BCGvaccination or immunostimulation against bladder cancer (1,21). Likewise, the panel could contribute to important publichealth investigations, as with M. bovis transmission from cattleto humans (27, 33). Since PCR is increasingly used, it is antic-ipated that both reference institutions and clinical diagnosticlaboratories may be able to adopt our strategy for MtbC sub-species determination.
In addition to providing a streamlined means to evaluateMtbC strain identity, this work expands upon the presentknowledge of genotypic features of the MtbC. First, Rv0577was evaluated and identified as an MtbC-restricted gene. Thecommonly used MtbC-defining element IS6110 has beenshown to be absent from some M. tuberculosis strains (39) andto be present in some MOTT strains (27). Hence, Rv0577 maybe a preferable MtbC PCR marker. Second, a novel 57-bpdeletion within Rv3879c was identified in three M. tuberculosisisolates. This new RD locus joins the previously describedRvD1 through -5 and TbD1 LSPs of numerous M. tuberculosisstrains (7) and suggests that an epidemiological relationshipexists among these three isolates. Together, the collective RDlocus data support the theory that the accumulation of LSPs isa significant adaptive strategy of the MtbC subspecies thatserves to generate genetic and biological diversity. Therefore,it is expected that other M. tuberculosis LSPs will be found andthat such LSPs will offer an additional means of assigninggenetic relatedness to M. tuberculosis strains. Third, the lociRD7 through RD10 were confirmed to be present in M. tuber-culosis, M. africanum subtype II, and “M. canettii” and con-firmed absent in the evaluated MtbC group 1a subspecies.Importantly, PCR amplification testing for these LSPs allowedthe segregation of M. africanum subtype I from M. africanumsubtype II, and together they offer a genotypic basis for theclassically described M. bovis-like versus M. tuberculosis-likephenotypic differences of M. africanum isolates (17). Recentstudies have also described additional M. africanum subtypeI-like strains that possess the RD7 locus but lack the RD9 locusand are unlike any strains evaluated in this report (7, 29, 35).These strains likely represent phylogenetic precursors of themore common M. africanum subtype I isolates but may alsoprove to be a novel MtbC subspecies. However, without exclu-
sive genetic markers for M. africanum subtype I, the taxonomicstatus of these unique strains containing RD7 but lacking RD9remains unclear. As a potential aid to type these strains, anRD9 primer pair was evaluated in this study and is included inthe list of primers in Table 1 (running at 600 bp, this ampliconfits well between lanes 6 and 7 in the PCR fragment cascadepresented in Fig. 1). Finally, the previously identified MtbCsubspecies- or lineage-specific SNPs of gyrA95, katG203,katG463, oxyR285, pncA57, hsp65631, and gyrB� were used col-lectively here for the first time and correlated perfectly withone another and with the RD locus data for all tested strainsexcept one. This M. africanum subtype II strain, 15082, is ofparticular interest because it had the appropriate 1234567 pro-file, contained RD9, and was MtbC genotypic group 1b (bykatG203, katG463, and gyrA95), but it bore the gyrBAf
(6446G3T SNP, M. africanum subtype I) sequence for thisamplicon. Until now, a gyrB� with the 6446G3T SNP has beendescribed only for MtbC genotypic group 1a strains (24, 30),thereby suggesting that strain 15082 (if the gyrB� 6446G3There is not simply a chance homologous mutation) is a remnantof the strain(s) that diverged from the preexisting MtbC lin-eage to become the group 1a MtbC subspecies (7). If so, thenthe gyrB� 6446G3T SNP arose prior to the fixation of thekatG203 SNP and also before the accumulation of the knownLSPs of the MtbC group 1a subspecies.
ACKNOWLEDGMENTS
We are indebted to Lee W. Riley, Sabine Ehrt, Walter Haas, GabyPfyffer, Barry Kreiswirth, Davise Larone, and Timothy Kiehn for pro-viding vital bacterial isolates. We also thank Hongxia Zhu, Petra deHaas, Krishna Menon, Benjamin See, and Susan Mossarella for theirtime and effort in maintaining and preparing isolates for our use. Weare also grateful to Barry Kreiswirth for critical appraisal of the manu-script and to Warren D. Johnson for trust and encouragement.
Funding support was provided by NIH grants R0-1 AI39606 andR0-1 HL61960 (to J.L.H.), an NIH Fogarty International CenterTraining grant (FICTG) (D43 TW00018), a grant from the Coorde-nacao de Aperfeicoamento de Pessoal de Nivel Superior (CAPES)(Ministry of Education-Brazil), and a grant from the Laura Cook HullTrust Fund (LCHTF) (Warren D. Johnson, principal investigator).R.C.H. was supported by the LCHTF, and L.C.D.O.L. was a FICTGand CAPES trainee. D.V.S. received funding from EC grant 200-0630for the Molecular Epidemiology of Tuberculosis.
ADDENDUM
After submission of this paper, M. africanum subtype IIstrain 15082 was PCR tested for the TbD1 locus. As describedrecently, this locus is believed to be absent from “modern” M.tuberculosis strains but present in both “ancient” M. tuberculo-sis strains and MtbC genotypic group 1a strains (6). Strain15082 yielded a DNA fragment for TbD1 by PCR that wasdetermined to be of the appropriate nucleotide sequence.These data are in keeping with our hypothesis that this strainis positioned along the MtbC phylogenetic tree intermediate tothe MtbC genotypic groups 1a and 1b.
REFERENCES
1. Anonymous. 1998. Case records of the Massachusetts General Hospital.Weekly clinicopathological exercises. Case 29–1998. A 57-year-old man withfever and jaundice after intravesical instillation of bacille Calmette-Guerinfor bladder cancer. N. Engl. J. Med. 339:831–837.
2. Aranaz, A., E. Liebana, E. Gomez-Mampaso, J. C. Galan, D. Cousins, A.Ortega, J. Blazquez, F. Baquero, A. Mateos, G. Suarez, and L. Dominguez.
1648 HUARD ET AL. J. CLIN. MICROBIOL.
1999. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomicstudy of a new member of the Mycobacterium tuberculosis complex isolatedfrom goats in Spain. Int. J. Syst. Bacteriol. 49:1263–1273.
3. Aranaz, A., E. Liebana, A. Mateos, L. Dominguez, D. Vidal, M. Domingo, O.Gonzolez, E. F. Rodriguez-Ferri, A. E. Bunschoten, J. D. Van Embden, andD. Cousins. 1996. Spacer oligonucleotide typing of Mycobacterium bovisstrains from cattle and other animals: a tool for studying epidemiology oftuberculosis. J. Clin. Microbiol. 34:2734–2740.
4. Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane,and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520–1523.
5. Brodin, P., K. Eiglmeier, M. Marmiesse, A. Billault, T. Garnier, S. Niemann,S. T. Cole, and R. Brosch. 2002. Bacterial artificial chromosome-based com-parative genomic analysis identifies Mycobacterium microti as a naturalESAT-6 deletion mutant. Infect. Immun. 70:5568–5578.
6. Brosch, R., S. V. Gordon, A. Billault, T. Garnier, K. Eiglmeier, C. Soravito,B. G. Barrell, and S. T. Cole. 1998. Use of a Mycobacterium tuberculosisH37Rv bacterial artificial chromosome library for genome mapping, se-quencing, and comparative genomics. Infect. Immun. 66:2221–2229.
7. Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K.Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Par-sons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A newevolutionary scenario for the Mycobacterium tuberculosis complex. Proc.Natl. Acad. Sci. USA 99:3684–3689.
8. Brunello, F., M. Ligozzi, E. Cristelli, S. Bonora, E. Tortoli, and R. Fontana.2001. Identification of 54 mycobacterial species by PCR-restriction fragmentlength polymorphism analysis of the hsp65 gene. J. Clin. Microbiol. 39:2799–2806.
9. Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V.Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D.Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T.Feltwell, S. Gentiles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jegels, A.Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail,M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares,S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998.Deciphering the biology of Mycobacterium tuberculosis from the completegenome sequence. Nature 393:537–544.
10. Eisenach, K. D., J. T. Crawford, and J. H. Bates. 1986. Genetic relatednessamong strains of the Mycobacterium tuberculosis complex. Analysis of restric-tion fragment heterogeneity using cloned DNA probes. Am. Rev. Respir.Dis. 133:1065–1068.
11. Frothingham, R., H. G. Hills, and K. H. Wilson. 1994. Extensive DNAsequence conservation throughout the Mycobacterium tuberculosis complex.J. Clin. Microbiol. 32:1639–1643.
12. Frothingham, R., P. L. Strickland, G. Bretzel, S. Ramaswamy, J. M. Musser,and D. L. Williams. 1999. Phenotypic and genotypic characterization ofMycobacterium africanum isolates from West Africa. J. Clin. Microbiol. 37:1921–1926.
13. Goh, K. S., E. Legrand, C. Sola, and N. Rastogi. 2001. Rapid differentiationof “Mycobacterium canettii” from other Mycobacterium tuberculosis complexorganisms by PCR-restriction analysis of the hsp65 gene. J. Clin. Microbiol.39:3705–3708.
14. Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T.Cole. 1999. Identification of variable regions in the genomes of tuberclebacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643–655.
15. Gordon, S. V., B. Heym, J. Parkhill, B. Barrell, and S. T. Cole. 1999. Newinsertion sequences and a novel repeated sequence in the genome of Myco-bacterium tuberculosis H37Rv. Microbiology 145:881–892.
16. Gutierrez, M., S. Samper, M. S. Jimenez, J. D. van Embden, J. F. Marin, andC. Martin. 1997. Identification by spoligotyping of a caprine genotype inMycobacterium bovis strains causing human tuberculosis. J. Clin. Microbiol.35:3328–3330.
17. Haas, W. H., G. Bretzel, B. Amthor, K. Schilke, G. Krommes, S. Rusch-Gerdes, V. Sticht-Groh, and H. J. Bremer. 1997. Comparison of DNA fin-gerprint patterns of isolates of Mycobacterium africanum from east and westAfrica. J. Clin. Microbiol. 35:663–666.
18. Haas, W. H., K. Schilke, J. Brand, B. Amthor, K. Weyer, P. B. Fourie, G.Bretzel, V. Sticht-Groh, and H. J. Bremer. 1997. Molecular analysis of katGgene mutations in strains of Mycobacterium tuberculosis complex from Africa.Antimicrob. Agents Chemother. 41:1601–1603.
19. Holland, S. M. 2001. Nontuberculous mycobacteria. Am. J. Med. Sci. 321:49–55.
20. Horstkotte, M. A., I. Sobottka, C. K. Schewe, P. Schafer, R. Laufs, S.Rusch-Gerdes, and S. Niemann. 2001. Mycobacterium microti llama-typeinfection presenting as pulmonary tuberculosis in a human immunodefi-ciency virus-positive patient. J. Clin. Microbiol. 39:406–407.
21. Jouanguy, E., F. Altare, S. Lamhamedi, P. Revy, J. F. Emile, M. Newport, M.Levin, S. Blanche, E. Seboun, A. Fischer, and J. L. Casanova. 1996. Inter-feron-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N. Engl. J. Med. 335:1956–1961.
22. Kallenius, G., T. Koivula, S. Ghebremichael, S. E. Hoffner, R. Norberg, E.
Svensson, F. Dias, B. I. Marklund, and S. B. Svenson. 1999. Evolution andclonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J. Clin.Microbiol. 37:3872–3878.
23. Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S.Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. vanEmbden. 1997. Simultaneous detection and strain differentiation of Myco-bacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol.35:907–914.
24. Kasai, H., T. Ezaki, and S. Harayama. 2000. Differentiation of phylogeneti-cally related slowly growing mycobacteria by their gyrB sequences. J. Clin.Microbiol. 38:301–308.
25. Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans,C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus,J. M. Musser, and J. D van Embden. 1999. Comparison of methods based ondifferent molecular epidemiological markers for typing of Mycobacteriumtuberculosis complex strains: interlaboratory study of discriminatory powerand reproducibility. J. Clin. Microbiol. 37:2607–2618.
26. Leclerc, M. C., N. Haddad, R. Moreau, and M. F. Thorel. 2000. Molecularcharacterization of environmental mycobacterium strains by PCR-restrictionfragment length polymorphism of hsp65 and by sequencing of hsp65, and of16S and ITS1 rDNA. Res. Microbiol. 151:629–638.
27. Liebana, E., A. Aranaz, B. Francis, and D. Cousins. 1996. Assessment ofgenetic markers for species differentiation within the Mycobacterium tuber-culosis complex. J. Clin. Microbiol. 34:933–938.
28. Mitarai, S., A. Kurashima, A. Tamura, H. Nagai, and H. Shishido. 2001.Clinical evaluation of Amplicor Mycobacterium detection system for thediagnosis of pulmonary mycobacterial infection using sputum. Tuberculosis81:319–325.
29. Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002.Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosiscomplex. J. Infect. Dis. 186:74–80.
30. Niemann, S., D. Harmsen, S. Rusch-Gerdes, and E. Richter. 2000. Differ-entiation of clinical Mycobacterium tuberculosis complex isolates by gyrBDNA sequence polymorphism analysis. J. Clin. Microbiol. 38:3231–3234.
31. Niemann, S., E. Richter, and S. Rusch-Gerdes. 2000. Differentiation amongmembers of the Mycobacterium tuberculosis complex by molecular and bio-chemical features: evidence for two pyrazinamide-susceptible subtypes of M.bovis. J. Clin. Microbiol. 38:152–157.
32. Niemann, S., S. Rusch-Gerdes, M. L. Joloba, C. C. Whalen, D. Guwatudde,J. J. Ellner, K. Eisenach, N. Fumokong, J. L. Johnson, T. Aisu, R. D.Mugerwa, A. Okwera, and S. K. Schwander. 2002. Mycobacterium africanumsubtype II is associated with two distinct genotypes and is a major cause ofhuman tuberculosis in Kampala, Uganda. J. Clin. Microbiol. 40:3398–3405.
33. O’Reilly, L. M., and C. J. Daborn. 1995. The epidemiology of Mycobacteriumbovis infections in animals and man: a review. Tuber. Lung Dis. 76(Suppl.1):1–46.
34. Park, H., H. Jang, C. Kim, B. Chung, C. L. Chang, S. K. Park, and S. Song.2000. Detection and identification of mycobacteria by amplification of theinternal transcribed spacer regions with genus- and species-specific PCRprimers. J. Clin. Microbiol. 38:4080–4085.
35. Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D.van Soolingen, Y. M. Hale, and M. Salfinger. 2002. Rapid and simple ap-proach for identification of Mycobacterium tuberculosis complex isolates byPCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339–2345.
36. Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazi-namidase/nicotinamidase, cause resistance to the antituberculous drug pyr-azinamide in tubercle bacillus. Nat. Med. 2:662–667.
37. Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Bottger.1996. Two-laboratory collaborative study on identification of mycobacteria:molecular versus phenotypic methods. J. Clin. Microbiol. 34:296–303.
38. Sreevatsan, S., P. Escalante, X. Pan, D. A. Gillies II, S. Siddiqui, C. N.Khalaf, B. N. Kreiswirth, P. Bifani, L. G. Adams, T. Ficht, V. S. Perumaalla,M. D. Cave, J. D. van Embden, and J. M. Musser. 1996. Identification of apolymorphic nucleotide in oxyR specific for Mycobacterium bovis. J. Clin.Microbiol. 34:2007–2010.
39. Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth,T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymor-phism in the Mycobacterium tuberculosis complex indicates evolutionarilyrecent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869–9874.
40. Talbot, E. A., D. L. Williams, and R. Frothingham. 1997. PCR identificationof Mycobacterium bovis BCG. J. Clin. Microbiol. 35:566–569.
41. Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity ofquality-controlled 16S rRNA gene sequence databases: identifying nontuber-culous Mycobacterium species. J. Clin. Microbiol. 39:3637–3648.
42. van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. van DerZeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin ofthe direct repeat locus of Mycobacterium tuberculosis complex bacteria. J.Bacteriol. 182:2393–2401.
43. van Soolingen, D., T. Hoogenboezem, P. E. de Haas, P. W. Hermans, M. A.Koedam, K. S. Teppema, P. J. Brennan, G. S. Besra, F. Portaels, J. Top,
VOL. 41, 2003 DIFFERENTIATION OF M. TUBERCULOSIS COMPLEX SUBSPECIES 1649
L. M. Schouls, and J. D. van Embden. 1997. A novel pathogenic taxon of theMycobacterium tuberculosis complex, Canetti: characterization of an excep-tional isolate from Africa. Int. J. Syst. Bacteriol. 47:1236–1245.
44. van Soolingen, D., A. G. van der Zanden, P. E. de Haas, G. T. Noordhoek, A.Kiers, N. A. Foudraine, F. Portaels, A. H. Kolk, K. Kremer, and J. D. vanEmbden. 1998. Diagnosis of Mycobacterium microti infections among hu-
mans by using novel genetic markers. J. Clin. Microbiol. 36:1840–1845.45. Viana-Niero, C., C. Gutierrez, C. Sola, I. Filliol, F. Boulahbal, V. Vincent,
and N. Rastogi. 2001. Genetic diversity of Mycobacterium africanum clinicalisolates based on IS6110-restriction fragment length polymorphism analysis,spoligotyping, and variable number of tandem DNA repeats. J. Clin. Micro-biol. 39:57–65.
1650 HUARD ET AL. J. CLIN. MICROBIOL.