Prefrontal-temporal gray matter deficits in bipolar disorder patients with persecutory delusions
ANK3 and CACNA1C – Missing genetic link for bipolar disorder and major depressive disorder in two...
-
Upload
independent -
Category
Documents
-
view
3 -
download
0
Transcript of ANK3 and CACNA1C – Missing genetic link for bipolar disorder and major depressive disorder in two...
at SciVerse ScienceDirect
Journal of Psychiatric Research 46 (2012) 973e979
Contents lists available
Journal of Psychiatric Research
journal homepage: www.elsevier .com/locate/psychires
ANK3 and CACNA1C e Missing genetic link for bipolar disorder and majordepressive disorder in two German case-control samples
Stefan Kloiber*, Darina Czamara, Nazanin Karbalai, Bertram Müller-Myhsok, Johannes Hennings,Florian Holsboer, Susanne LucaeMax Planck Institute of Psychiatry, Kraepelinstrasse 2-10, Munich, Germany
a r t i c l e i n f o
Article history:Received 15 February 2012Received in revised form16 April 2012Accepted 19 April 2012
Keywords:DepressionGeneticsBipolar disorderCACNA1CANK3
* Corresponding author. Tel.: þ49 89306220; fax: þE-mail address: [email protected] (S. Klo
0022-3956/$ e see front matter � 2012 Elsevier Ltd.doi:10.1016/j.jpsychires.2012.04.017
a b s t r a c t
Recent genome-wide association studies (GWAS) and metaanalyses revealed genetic associations forANK3 (ankyrin 3) and CACNA1C (alpha 1C subunit of the L-type voltage gated calcium channel) withbipolar disorder (BPD). Several findings from clinical, epidemiological, and genetic studies point towardsa common biological background of BPD and major depressive disorder (MDD). We were interestedwhether this also applies for ANK3 and CACNA1C and tested associations of single nucleotide poly-morphisms (SNPs) in these genes with MDD in two Caucasian case-control samples. Sample 1 (MunichAntidepressant Response Signature Project/MARS e MDD) consisted of 720 depressed inpatients and542 psychiatric healthy controls. Sample 2 (unipolar recurrent depression (URD)) consisted of 827patients with URD and 860 psychiatric healthy controls. After stringent quality control we analyzed 262SNPs (sample 1) and 504 SNPs (sample 2) and imputed further 5771 SNPs (sample 1) and 5534 SNPs(sample 2) from Hapmap Phase 2 data in the ANK3 and CACNA1C gene regions. Additionally, a meta-analysis of both samples was performed. Several SNPs in both genes were nominally associated withMDD with the highest association in the 30-region of ANK3 (rs10994143, nominal p ¼ 3.3*10�4) in themetaanalysis of both samples. None of these results remained significant after correction for multipletesting. No association of MDD with SNPs previously reported in BPD studies could be detected. Byanalyzing the LD-structure, our highest associated SNPs could not be linked to the SNPs previouslyreported in BPD. Regarding ANK3 and CACNA1C, our findings do not support a strong genetic linkbetween BPD and MDD for these two genes.
� 2012 Elsevier Ltd. All rights reserved.
1. Introduction
From recent genome-wide association studies (GWAS), replica-tion studies, and metaanalyses in bipolar disorder (BPD) (Chenet al., 2011; Ferreira et al., 2008; Schulze et al., 2008; Scott et al.,2009; Sklar et al., 2011, 2008; Tesli et al., 2011; Wray et al., 2012)two genes, ANK3 (ankyrin 3) and CACNA1C (alpha 1C subunit of theL-type voltage gated calcium channel), have been reported to beimplicated in disease aetiology. The association of ANK3 poly-morphisms with BPD was additionally confirmed in two Asiansamples (Lee et al., 2011; Takata et al., 2011). One smaller associa-tion study did not support this association (Weber et al., 2011).
Furthermore, cross-disorder analyses of schizophrenia and BPDsamples (Athanasiu et al., 2010; Green et al., 2009; Ripke et al.,2011; Sklar et al., 2011; Tesli et al., 2011; Williams et al., 2011)
49 8930622605.iber).
All rights reserved.
predominantly supported a potential relevance of these genes inpsychiatric phenotypes.
So far, the cross-disorder relevance of ANK3 and CACNA1C forBPD and major depressive disorder (MDD) has not been investi-gated as extensively. Green et al. (2009) described an association ofrs1006737, a polymorphism in CACNA1C detected in the BPDstudies, with MDD. In a recent metaanalysis (Liu et al., 2011)a cross-disorder association for BPD and MDD could be confirmedfor CACNA1C, whereas ANK3 polymorphisms were not associatedin this BPD e MDD metaanalysis.
Mutations in calcium channel genes (L-type and T-type, CAC-NA1H) resulting in altered calcium signalling have been previouslyshown to be involved in autism spectrum disorders (Splawski et al.,2004, 2006). The close relationship of calcium channels, calmod-ulin and calcium/calmodulin dependent pathways and their role insynaptic plasticity via regulation of cytoskeletal structure forma-tion (Xia and Storm, 2005) could be a possible explanation for theinvolvement of calcium channels in psychiatric disorders. Ankyrinshave been suggested to be involved in the organization of neuronal
S. Kloiber et al. / Journal of Psychiatric Research 46 (2012) 973e979974
subcellular structure and synapse formation (Huang, 2006). ANK3polymorphisms have been linked to decreased white matterintegrity (Linke et al., 2012) and ANK3 has been suggested to playan important role in neurogenesis (Paez-Gonzalez et al., 2011).Furthermore, preliminary results on ankyrin dependent membranealterations in bipolar patients (Meltzer, 1991; Zhang and Meltzer,1989) underscore a potential role of these cytoskeletal proteins indifferent brain disorders.
Besides ANK3 and CACNA1C, a putatively shared geneticsusceptibility locus for BPD and MDD located at chromosome3p21.1 has heated up the discussion about GWAS cross-disordermetaanalyses challenging traditional diagnostic boundaries ofpsychiatric disorders (Breen et al., 2010; McMahon et al., 2010a,b).Additionally, genetic association studies in BPD and MDD pointedtowards candidate genes associated with the susceptibility for bothdisorders, for example TPH2 (Cichon et al., 2007; Van Den Bogaertet al., 2006; Zhang et al., 2005) and P2RX7 (Barden et al., 2006;Lucae et al., 2006).
An extensive discussion about BPD and MDD as related disor-ders or different disease entities has been ongoing for decades(Cassano et al., 2004; Mondimore, 2005; Winokur et al., 1995).Differences in psychopathology with manic and hypomanicepisodes in BPD, in disease course like earlier age of onset andhigher episode frequency in bipolar patients (Winokur et al., 1993),as well as differences in clinical features like more frequent retar-dation and psychotic symptoms in BPD (Mondimore, 2005) haveled to consider MDD and BPD as distinct disease entities (Winokuret al., 1995). However, the indistinguishable psychopathologicalsymptoms of patients with recurrent MDD and BPD duringdepressive episodes strongly suggest that both clinical conditionsshare a common set of causal factors. Furthermore, twin studiesrevealed a genetic overlap of MDD and BPD (McGuffin et al., 2003)and suggested common genetic factors for both diseases. Familystudies led to the continuum hypothesis of MDD, BPD, and schiz-oaffective disorder (Gershon et al., 1982) and pointed to a familialaggregation of MDD in relatives of patients suffering from BPD(Andreasen et al., 1987; McGuffin and Katz, 1989; Winokur et al.,1995). The identification of manic and hypomanic symptoms inpatients with MDD (Cassano et al., 2004) challenges the concept ofan unipolar-bipolar dichotomy. Several large long-term studiesexamining the course of mood disorders reported a substantial rateof patients with a single major depressive episode or MDD devel-oping BPD in a later phase of life (Akiskal et al., 1995; Angst et al.,2005; Mondimore, 2005) providing evidence for MDD and BPD asa common disease entity.
These findings that favor the hypothesis of a common set ofpathological mechanisms accounting for BPD and MDD led us toinvestigate whether the recently identified genes, ANK3 and CAC-NA1C, associated with BPD (Ferreira et al., 2008) are associatedwith MDD in our two depression samples. GWAS data of thesesamples have been previously reported (Kohli et al., 2011; Mugliaet al., 2008).
2. Materials and methods
2.1. Sample description
Subjects with available genome-wide genotyping data wereselected for analysis from both samples.
2.1.1. Sample 1 (Munich Antidepressant Response Signature Project/MARS-MDD)
720 depressive in patients (336 males, 384 females) wererecruited for the Munich Antidepressant Response Signature(MARS) project at the Max Planck Institute of Psychiatry in Munich,
Germany (www.mars-depression.de). The mean age (�SD) was48.68 � 14.13 years. Study details have been reported by Henningset al. (2009) and a genome-wide association study has been pub-lished previously (Kohli et al., 2011). Patients suffered from a singledepressive episode or from a depressive episode occurring in thecourse of recurrent MDD (n ¼ 640) or from a depressive episodeoccurring in the course of BPD (n ¼ 80). All included patients wereof European descent and 94% were of German origin. 542 psychi-atric healthy control subjects (244 males, 298 females) werematched to the patient sample for age, gender, and ethnicity froma randomly selected Munich-based community sample. Theyunderwent computer-assisted interviews based on the Munichversion of the Composite International Diagnostic Interview (M-CIDI) (Wittchen et al., 1999). Exclusion criteria were lifetime historyof mood disorders, schizophrenia, anxiety disorders, alcoholdependence, drug abuse, obsessive-compulsive disorder, PTSD,dissociative disorders, somatoform disorders, and eating disorders.More detailed demographic and clinical characteristics of sample 1are presented in Supplementary Table 1.
2.1.2. Sample 2 (unipolar recurrent depression (URD))This independent sample consisted of 827 patients with URD
(278 males, 549 females). The mean age (�SD) was 50.90 � 13.72years. 860 psychiatric healthy control subjects (278 males, 582females) were matched to the patient sample for age, gender, andethnicity from a randomly selected Munich-based communitysample. Study details have been reported by Lucae et al. (2006) anda genome-wide association study has been published previously(Muglia et al., 2008). More detailed demographic and clinicalcharacteristics of sample 2 are presented in Supplementary Table 1.
2.2. DNA preparation
On enrolment, 40 ml of EDTA blood were drawn from eachsubject. DNA was extracted from fresh blood using the Puregenewhole blood DNA extraction kit (Gentra Systems Inc., MN, USA).
2.3. SNP genotyping, quality control, imputation, and statisticalanalysis
Sample 1: Genotyping was performed with HumanHap100 andHumanHap300 BeadChips (Illumina Inc., San Diego, California,USA) genome-wide genotyping arrays according to the standardprotocol of the manufacturer.
Sample 2: Genotyping was performed by HumanHap 550BeadChips (Illumina Inc., San Diego, California, USA) genome-widegenotyping arrays according to the standard protocol of themanufacturer.
As sample 1 contained patients with BPD, we performed twoanalyses including and excluding the bipolar patients.
We included single nucleotide polymorphisms (SNPs)located � 150 kb from gene borders by selecting the longest iso-forms according to UCSC Genome Browser hg18; www.genome.ucsc.edu: ANK3 gene region: (chr10:61,306,062-62,313,290), CAC-NA1C gene region: (chr12:1,882,677-2,827,376).
Imputation was performed using impute2 (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html). Missing genotypes wereestimated based on the linkage disequilibrium (LD) structurebetween markers in the reference samples from HapMap3 (CEU-individuals, release #2 from Feb 2009) and the 1000 Genomesproject (CEU-individuals, data freeze from March 2010). Prior toimputation, we ran a strict quality control (QC) on genotyped datausing only SNPs with callrates of at least 0.98 and excluding SNPs
S. Kloiber et al. / Journal of Psychiatric Research 46 (2012) 973e979 975
with a p-value for deviation from Hardy-Weinberg-Equilibrium(HWE; Wigginton et al., 2005) of lower than 1.0*10�5. After QC,128 genotyped SNPs located in the ANK3 region and 134 genotypedSNPs located in the CACNA1C region were available from sample 1.252 genotyped SNPs in the ANK3 region and 252 genotyped SNPs inthe CACNA1C region from sample 2, respectively. For sample 1, 3155SNPs in the ANK3 region and 2616 SNPs in the CACNA1C region, forsample 2, 3034 SNPs in the ANK3 region and 2500 SNPs in theCACNA1C region were imputed.
Allelic association analysis was performed using PLINK (http://pngu.mgh.harvard.edu/wpurcell/plink/) and theedosage option.SNPswith aminorallele frequencybelow0.01 aswell as SNPswith animputation info scorebelow0.9orhigher than1 (implicatingpossibledeviation from HWE) were excluded from statistical analysis.
After this second quality control, 1684 SNPs in sample 1including patients with BPD (ANK3), 1620 SNPs in sample 1excluding patients with BPD (ANK3), 854 SNPs in sample 1including patients with BPD (CACNA1C), 824 SNPs in sample 1excluding patients with BPD (CACNA1C), 1819 SNPs in sample 2(ANK3), and 972 SNPs in sample 2 (CACNA1C) remained for dataanalysis. A test for population stratification with 10,000 randomSNPs as genomic controls showed no evidence for admixture (Isinget al., 2009). Metaanalysis was performed using theemetaanalysisoption in PLINK under a fixed-effects model. As no populationadmixture was detected, an additional association test was per-formed on both samples in a combined study containing all indi-viduals from both samples.
Table 1aBest association results for ANK3 and CACNA1C polymorphisms in sample 1, sample 2, and(protective allele) more frequent in controls. OR: odds ratio. CI: confidence interval.
SNP Chr Position Allele 1 Allele 2 Freq allele 1
ANK3Sample 1rs12265962 10 61457557 G T 0.014rs10994133 10 61435413 C T 0.99rs10994143 10 61451084 A G 0.99rs3793862 10 61615065 T C 0.94rs12355908 10 61607010 C T 0.74Sample 2rs10821634 10 61419849 A C 0.82rs55960897 10 61421215 T C 0.82rs10821635 10 61420539 T A 0.82rs10761429 10 61422206 T A 0.82rs10821636 10 61422061 A G 0.82Metaanalysis Sample1rs10994143 10 61451084 A G 0.99rs12265962 10 61457557 G T 0.014rs10821700 10 61629067 C A 0.82rs7908312 10 61667891 A C 0.83rs10821707 10 61667962 A G 0.83
CACNA1CSample 1rs2238071 12 2326677 A G 0.42rs2238070 12 2326376 G T 0.43rs3819534 12 2307129 A G 0.39rs2283302 12 2322880 A G 0.30rs3819532 12 2307098 T C 0.40Sample 2rs11062247 12 2486389 A G 0.80rs4765681 12 2427457 T C 0.43rs16929470 12 2472003 C T 0.95rs11062248 12 2486449 A T 0.80rs2239073 12 2408761 T C 0.56Metaanalysis Sample1rs2239073 12 2408761 T C 0.56rs2238065 12 2312892 A G 0.28rs2238066 12 2315660 G A 0.29rs4765937 12 2440796 T C 0.56rs38119536 12 2307259 G A 0.31
To correct formultiple testing theBonferronimethodwas applied.To consider a less conservative method as Bonferroni, we performeda permutation based method for multiple testing with 100,000permutations as described by Westfall and Young (W/Y) (1993) totake advantage of the dependence structure between SNPs.
For examination of LD patterns we analyzed r2 measures(Hill and Robertson, 1968) using PLINK in control individuals ofsample 1.
To estimate the probability of false negative results whenanalyzing polymorphisms previously shown to be associated withBPD we performed a power analysis using QUANTO, Version 1.2.4,May 2009 (http://hydra.usc.edu/gxe/). The power to detectcomparable results as described in the Ferreira et al. (2008) studywith a supposed population risk for MDD of 0.15 was >0.6 forsample 1, > 0.7 sample 2, and >0.9 for both samples.
3. Results
3.1. ANK3
In sample 1 and sample 2 the lowest nominal p-values could bedetected for rs12265962 and rs10821634, respectively (Table 1a).In the metaanalysis (sample 1 and sample 2) the best nominal p-value (3.30*10�4) was observed for rs10994143 (Table 1a).Although, this SNP comprised a low minor allele frequency,genotype distribution in both samples was highly consistent:Sample 1: patients: AA n ¼ 703, AG n ¼ 9, GG n ¼ 0; controls: AA
metaanalysis. OR> 1: allele 1 (risk allele) more frequent in patients. OR< 1: allele 1
OR (CI) p-value
0.41 (0.30e0.56) 0.0212.39 (0.40e14.28) 0.0222.38 (0.40e14.02) 0.0231.53 (0.85e2.76) 0.0271.25 (0.97e1.61) 0.027
1.31 (1.04e1.66) 0.00271.31 (1.04e1.66) 0.00291.32 (1.04e1.67) 0.00301.31 (1.04e1.65) 0.00311.31 (1.04e1.65) 0.0031
Sample 2 Sample1 Sample 20.98 2.38 (0.40e14.02) 2.11 (0.70e6.37) 0.000330.017 0.41 (0.30e0.56) 0.48 (0.37e0.62) 0.000370.83 0.84 (0.69e1.02) 0.80 (0.69e0.80) 0.00530.83 0.87 (0.71e1.07) 0.79 (0.69e0.92) 0.00700.83 0.87 (0.71e1.06) 0.80 (0.69e0.92) 0.0075
1.34 (1.06e1.69) 0.000881.33 (1.05e1.66) 0.00131.32 (1.05e1.66) 0.00191.35 (1.04e1.76) 0.00201.32 (1.04e1.66) 0.0025
0.79 (0.68e0.89) 0.00480.82 (0.73e0.92) 0.00540.65 (0.53e0.79) 0.00550.78 (0.68e0.90) 0.00590.83 (0.74e0.94) 0.010
Sample 2 Sample1 Sample 20.55 0.95 (0.80e1.12) 0.83 (0.74e0.94) 0.0170.28 1.33 (1.02e1.72) 1.06 (0.90e1.25) 0.0210.29 1.33 (1.04e1.71) 1.02 (0.88e1.19) 0.0400.29 0.92 (0.78e1.09) 0.88 (0.77e0.99) 0.0430.30 1027 (1.00-1.61) 1.04 (0.89-1.22) 0.047
S. Kloiber et al. / Journal of Psychiatric Research 46 (2012) 973e979976
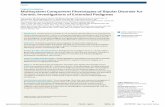
n ¼ 520, AG n ¼ 16, GG n ¼ 1; Sample 2: patients: AA n ¼ 809, AGn ¼ 14, GG n ¼ 0; controls: AA n ¼ 821, AG n ¼ 35, GG n ¼ 1. Aftercorrection for multiple testing with Bonferroni and W/Y methodsresults did not remain significant at the significance-level of 0.05,�log p-values of metaanalysis results are plotted in Fig. 1 anddetailed results for all polymorphisms in ANK3 for sample 1,
Fig. 1. Metaanalysis (sample 1 and sample 2) association results of genotyped and imputed�log10 p-values are shown.
sample 2, and metaanalysis are available in SupplementaryTables 2, 3, and 4.
SNPs previously reported from GWAS for BPD (Ferreira et al.,2008; Schulze et al., 2008) (rs10994336, rs1938526, rs9804190)and combined analysis for schizophrenia and BPD (Ripke et al.,2011) (rs10994359) were preferentially located upstream of
SNPs in the ANK3 region on chromosome 10 and CACNA1C region on chromosome 12.
S. Kloiber et al. / Journal of Psychiatric Research 46 (2012) 973e979 977
polymorphisms showing the best association results in our meta-analysis. We could not detect significant association results forthese previously reported polymorphisms in our samples andmetaanalysis (Table 1b). Minimum and maximum distancebetween these previously reported polymorphisms and SNPsshowing the best association results in ourmetaanalysis were 58 kband 519 kb, respectively. The most highly associated poly-morphisms in our metaanalysis and previously reported poly-morphisms from bipolar GWAS studies were not in LD (r2 < 0.1).
3.2. CACNA1C
In sample 1 and 2 the lowest nominal p-values could be detectedfor rs2238071 and rs11062247, respectively (Table 1a). The best p-value (0.017) in the metaanalysis was calculated for rs2239073(Table 1a). These results did not remain significant at thesignificance-level of 0.05 after correction for multiple testing withBonferroni andW/Ymethods,�log p-values of metaanalysis resultsare plotted in Fig. 1 and detailed results for all polymorphisms inCACNA1C for sample 1, sample 2, and metaanalysis are available inSupplementary Tables 5, 6, and 7.
We could not detect significant association results for previouslyreported polymorphisms reported from GWAS for BPD (Ferreiraet al., 2008; Sklar et al., 2011, 2008) (rs1006737, rs4765913,rs10848632) and combined analysis for schizophrenia and BPD(Ripke et al., 2011) (rs4765905) (Table 1b). These polymorphismswere located upstream of SNPs showing the best association resultsin our metaanalysis. Minimum and maximum distance betweenthese previously reported polymorphisms and polymorphismsshowing the best results in ourmetaanalysis were 17 kb and 150 kb,respectively. No LD between previously reported polymorphisms inbipolar GWAS studies and the most highly associated poly-morphisms in our metaanalysis could be detected (r2 < 0.2). Asthree SNPs from the BPD studies (rs4765913, rs1006737,rs10848632) were excluded due to quality control criteria insample 2, we searched for polymorphisms in high LD (r2 > 0.95) toestimate the results for these SNPs in sample 2. No significantresults for these proxy SNPs in sample 2 and metaanalysis could bedetected (rs4765913e rs10774037: r2> 0.99, alleles G/A: frequencyG 0.18, sample 2: OR ¼ 0.97, p ¼ 0.80; metaanalysis: OR ¼ 1.01,p ¼ 0.85; rs1006737 e rs4765905: r2 ¼ 1, results for rs4765905 arepresented in Table 1b; rs10848632 e rs55961529: r2 > 0.95, allelesG/A: frequency G 0.64, sample 2: OR ¼ 1.1, p ¼ 0.19; metaanalysis:OR ¼ 1.05, p ¼ 0.43).
3.3. Supplementary analyses
As described above, we performed additional analysis andmetaanalysis excluding patients with BPD from sample 1 (n ¼ 80),
Table 1bAssociation results for previously reported polymorphisms with genome-wide significanallele) more frequent in patients. OR < 1: allele 1 (protective allele) more frequent in co
SNP Chr Position Allele 1 Allele 2 Freq allele 1 Sample 1
OR (CI)
ANK3rs10994359 10 61892113 T C 0.93 1.21 (0.82e1rs10994336 10 61849818 C T 0.93 1.20 (0.81e1rs1938526 10 61970389 A G 0.92 1.20 (0.82e1rs9804190 10 61509837 C T 0.78 1.16 (0.91e1CACNA1Crs4765905 12 2219845 G C 0.67 0.92 (0.77e1rs4765913 12 2290157 A T 0.22 1.11 (0.87e1rs1006737 12 2215556 G A 0.67 0.9 (0.77e1rs10848632 12 2186254 C T 0.63 0.96 (0.81e1
which are presented in Supplementary Tables 8, 9, 10 and 11. Theseresults showed no or not more than marginal differences comparedto the results presented above using the entire sample 1.
Besides, we performed association analysis combining individ-uals from both samples. The results from this combined samplewere comparable to the metaanalyses results.
No significant results could be detected after correction formultiple testing in these supplementary analyses.
4. Discussion
We were not able to detect any significant associations of gen-otyped and imputed polymorphisms in the ANK3 and CACNA1Cgene regions with MDD after correction for multiple testing. Thelowest nominal p-values (10�4) were detected for polymorphismsin the 30-area of ANK3. These polymorphisms were not in LD andcould therefore not be linked with previously reported associatedpolymorphisms in BPD and in a large combined schizophrenia andBPD analysis.
Regarding the indistinguishable clinical presentation ofdepressive episodes in MDD and BPD and the high transmission ofboth disorders within families (Andreasen et al., 1987; Gershonet al., 1982; McGuffin and Katz, 1989; McGuffin et al., 2003;Winokur et al., 1995) the finding of genetic associations for bothdisorders in the same genes appears possible. Although epidemi-ological and clinical findings point towards a biological relationshipand common pathomechanisms of both BPD and MDD, our resultsfrom a large number of genotyped and imputed polymorphisms donot support the currently best validated genetic association resultsin BPD (CACNA1C or ANK3) as linking genetic factors. A previousstudy by Green et al. (2009) described an association of the CAC-NA1C polymorphism rs1006737 with MDD, when testing thispolymorphism exclusively. Furthermore, a metaanalysis of BPD andMDD samples supported a cross-disorder association with CAC-NA1C but not ANK3 polymorphisms (Liu et al., 2011). Additionally,the previously described association findings for other diseases, e.g.autism spectrum disorders (Splawski et al., 2004, 2006) andschizophrenia (Athanasiu et al., 2010; Green et al., 2009; Ripkeet al., 2011; Sklar et al., 2011; Tesli et al., 2011; Williams et al.,2011) point towards a participation of CACNA1C in psychiatricdisorders.
Besides these two genes (ANK3 and CACNA1C), several studiesdetected genetic loci pointing towards common susceptibility forBPD and MDD (Barden et al., 2006; Breen et al., 2010; Cichon et al.,2007; Lucae et al., 2006; McMahon et al., 2010a,b; Van Den Bogaertet al., 2006; Zhang et al., 2005).
In the previously published GWAS results from our two samples(Kohli et al., 2011; Muglia et al., 2008) polymorphisms located inthe ANK3 and CACNA1C regions did not range within the highest
ce in BPD. * SNPs were excluded due to quality control criteria. OR > 1: allele 1 (riskntrols.
Sample 2 Metaanalysis sample 1 and 2
p-value OR (CI) p-value OR (CI) p-value
.81) 0.24 1.20 (0.88e1.64) 0.17 1.21 (0.94e1.54) 0.07
.78) 0.29 1.17 (0.86e1.59) 0.24 1.19 (0.93e1.51) 0.11
.76) 0.25 1.21 (0.89e1.64) 0.15 1.20 (0.95e1.53) 0.07
.48) 0.16 1.06 (0.89e1.25) 0.50 1.09 (0.95e1.26) 0.16
.08) 0.35 1.09 (0.93e1.28) 0.22 1.02 (0.91e1.15) 0.69
.41) 0.32 NA* NA* NA* NA*
.07) 0.31 NA* NA* NA* NA*
.43) 0.69 NA* NA* NA* NA*
S. Kloiber et al. / Journal of Psychiatric Research 46 (2012) 973e979978
significant results. The best GWAS result in sample 1 was detectedon chromosome 12 with a distance of >80 Mb from the CACNA1Cregion. The closest SNPs among the best results in sample 2 werelocated at a distance of 2 Mb from the CACNA1C region and 24 Mbfrom the ANK3 region.
Although we analyzed a total of 2949 subjects, the sample size forgenetic associationanalysis is still small.However, the statistical powerinouranalyseswouldhavebeen sufficient todetect association results,e.g. as described by Ferreira et al. (2008). Regarding the ongoingdiscussion about potential false positive findings depending on allelefrequency (Tabangin et al., 2009), we would like to mention that theSNPs showing the highest association results in our analyses(rs10994143 and rs12265962) had low minor allele frequenciesbetween 0.01 and 0.02. However, the genotype distribution of thesepolymorphisms in both samples was highly consistent.
From aforementioned biological mechanisms of ANK3 and CAC-NA1C several findings point towards an involvement of ankyrins andcalcium channels in cytoskeletal processes of neuronal and synapticplasticity (Huang, 2006; Linke et al., 2012; Meltzer, 1991; Paez-Gonzalez et al., 2011; Splawski et al., 2004, 2006; Xia and Storm,2005; Zhang and Meltzer, 1989). From these findings and cross-disorder genetic studies listed above distinctions regarding the rele-vance of genes or mechanisms in different psychiatric disorders oraffective disorder subentities would be premature. Thus, further eval-uation of these genetic associations in multiple samples as well asfunctional studies in both disorders are necessary to be able toconverge to biological and measurable factors for distinguishing sub-entities of affective disorders (APA,1994; WHO, 2007).
Common as well as different genetic findings in MDD and BPDand findings from cross-disorder genetic studies are important andcould provide insights into common or different pathomechanismsof bothMDD and BPD. This could help to conceive the developmentof BPD in patients with recurrent MDD and could possibly challengethe diagnostic concepts of affective disorders.
Role of funding source
This work was funded by the Max Planck Society and in part bya research grant from the German Federal Ministry for Educationand Research (BMBF) in the framework of the National GenomeResearch Network (NGFN2 and NGFN-Plus, FKZ 01GS0481) and bythe BMBF Program “Molecular Diagnostics: Validation ofBiomarkers for Diagnosis and Outcome in Major Depression”(01ES0811). The authors’ research on personalized medicine issupported by the Max Planck Excellence Foundation.
Contributors
Stefan Kloiber wrote the first draft of the manuscript. Darina Cza-mara, Nazanin Karbalai, and Bertram Müller-Myhsok undertook thestatistical analysis and wrote parts of the manuscript. Stefan Kloiber,Johannes Hennings, and Susanne Lucae conducted the clinical studies.Florian Holsboer and Susanne Lucae designed the studies. All authorscontributed to and have approved the final manuscript.
Conflict of interest
The authors report no conflicts of interest.
Acknowledgements
We thank Manfred Uhr and Thomas Bettecken for supervisingthe laboratory and genotyping procedures and Gertrud Ernst-Jan-sen, Elisabeth Kappelmann, and Beate Siegel for excellent technicalassistance.
Appendix A. Supplementary material
Supplementary material related to this article can be foundonline at doi:10.1016/j.jpsychires.2012.04.017
References
Akiskal HS, Maser JD, Zeller PJ, Endicott J, Coryell W, Keller M, et al. Switching fromunipolar to bipolar-Ii e an 11-year prospective-study of clinical and tempera-mental predictors in 559 patients. Archives of General Psychiatry 1995;52:114e23.
Andreasen NC, Rice J, Endicott J, Coryell W, Grove WM, Reich T. Familial rates ofaffective-disorder e a report from the National-Institute-Of-Mental-Health-Collaborative-Study. Archives of General Psychiatry 1987;44:461e9.
Angst J, Sellaro R, Stassen HH, Gamma A. Diagnostic conversion from depression tobipolar disorders: results of a long-term prospective study of hospital admis-sions. Journal of Affective Disorders 2005;84:149e57.
APA. Diagnostic and statistical manual of mental disorders. 4th ed. Washington DC:American Psychiatric Association; 1994.
Athanasiu L, Mattingsdal M, Kähler AK, Brown A, Gustafsson O, Agartz I, et al.Gene variants associated with schizophrenia in a Norwegian genome-widestudy are replicated in a large European cohort. Journal of PsychiatricResearch 2010;44:748e53.
Barden N, Harvey M, Gagne B, Shink E, Tremblay M, Raymond C, et al. Analysis ofsingle nucleotide polymorphisms in genes in the chromosome 12Q24.31 regionpoints to P2RX7 as a susceptibility gene to bipolar affective disorder. AmericanJournal of Medical Genetics Part B: Neuropsychiatric Genetics 2006;141B:374e82.
Breen G, Lewis CM, Vassos E, Pergadia ML, Blackwood DHR, Boomsma DI, et al.Replication of association of 3p21.1 with susceptibility to bipolar disorder butnot major depression. Nature Genetics 2010;43:3e5.
Cassano GB, Rucci P, Frank E, Fagiolini A, Dell’Osso L, Shear MK, et al. The moodspectrum in unipolar and bipolar disorder: arguments for a unitary approach.American Journal of Psychiatry 2004;161:1264e9.
Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJM, et al. Genome-wideassociation study meta-analysis of European and Asian-ancestry samplesidentifies three novel loci associated with bipolar disorder. Molecular Psychi-atry; 2011.
Cichon S, Winge I, Mattheisen M, Georgi A, Karpushova A, Freudenberg J, et al.Brain-specific tryptophan hydroxylase 2 (TPH2): a functional Pro206Sersubstitution and variation in the 5’-region are associated with bipolar affectivedisorder. Human Molecular Genetics; 2007. ddm286.
Ferreira MAR, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al.Collaborative genome-wide association analysis supports a role for ANK3 andCACNA1C in bipolar disorder. Nature Genetics 2008;40:1056.
Gershon ES, Hamovit J, Guroff JJ, Dibble E, Leckman JF, Sceery W, et al. A Familystudy of schizoaffective, bipolar-I, bipolar-Ii, unipolar, and normal controlprobands. Archives of General Psychiatry 1982;39:1157e67.
Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S, et al. The bipolar disorderrisk allele at CACNA1C also confers risk of recurrent major depression and ofschizophrenia. Molecular Psychiatry; 2009.
Hennings JM, Owashi T, Binder EB, Horstmann S, Menke A, Kloiber S, et al. Clinicalcharacteristics and treatment outcome in a representative sample of depressedinpatients e findings from the Munich Antidepressant Response Signature(MARS) project. Journal of Psychiatric Research 2009;43:215e29.
Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theoretical andApplied Genetics 1968;38:226e31.
Huang ZJ. Subcellular organization of GABAergic synapses: role of ankyrins and L1cell adhesion molecules. Nature Neuroscience 2006;9:163.
Ising M, Lucae S, Binder EB, Bettecken T, Uhr M, Ripke S, et al. A genomewideassociation study points to multiple loci that predict antidepressant drugtreatment outcome in depression. Archives of General Psychiatry 2009;66:966e75.
Kohli Martin A, Lucae S, Saemann Philipp G, Schmidt Mathias V, Demirkan A, Hek K,et al. The neuronal transporter gene SLC6A15 confers risk to major depression.Neuron 2011;70:252e65.
Lee MTM, Chen CH, Lee CS, Chen CC, Chong MY, Ouyang WC, et al. Genome-wideassociation study of bipolar I disorder in the Han Chinese population. MolecularPsychiatry 2011;16:548e56.
Linke J, Witt SH, King AV, Nieratschker V, Poupon C, Gass A, et al. Genome-widesupported risk variant for bipolar disorder alters anatomical connectivity in thehuman brain. Neuroimage 2012;59:3288e96.
Liu Y, Blackwood DH, Caesar S, de Geus EJC, Farmer A, Ferreira MAR, et al. Meta-analysis of genome-wide association data of bipolar disorder and majordepressive disorder. Molecular Psychiatry 2011;16:2e4.
Lucae S, Salyakina D, Barden N, Harvey M, Gagne B, Labbe M, et al. P2RX7, a genecoding for a purinergic ligand-gated ion channel, is associated with majordepressive disorder. Human Molecular Genetics 2006;15:2438e45.
McGuffin P, Katz R. The genetics of depression and manic-depressive disorder.British Journal of Psychiatry 1989;155:294e304.
McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The Heritability ofbipolar affective disorder and the genetic relationship to unipolar depression.Archives of General Psychiatry 2003;60:497e502.
S. Kloiber et al. / Journal of Psychiatric Research 46 (2012) 973e979 979
McMahon FJ, Akula N, Cichon S, Detera-Wadleigh SD, Edenberg H, Holsboer F, et al.Reply to [ldquo]Replication of association of 3p21.1 with susceptibility tobipolar disorder but not major depression[rdquo]. Nature Genetics 2010a;43:5.
McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD, et al.Meta-analysis of genome-wide association data identifies a risk locus for majormood disorders on 3p21.1. Nature Genetics 2010b;42:128e31.
Meltzer HL. Is there a specific membrane defect in bipolar disorders. BiologicalPsychiatry 1991;30:1071e4.
Mondimore FM. Unipolar depression/bipolar depression: connections and contro-versies. International Review of Psychiatry 2005;17:39e47.
Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ, et al. Genome-wideassociation study of recurrent major depressive disorder in two European case-control cohorts. Molecular Psychiatry; 2008.
Paez-Gonzalez P, Abdi K, Luciano D, Liu Y, Soriano-Navarro M, Rawlins E, et al.Ank3-dependent SVZ niche assembly is required for the continued productionof new neurons. Neuron 2011;71:61e75.
Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nature Genetics2011;43:969e76.
Schulze TG, Detera-Wadleigh SD, Akula N, Gupta A, Kassem L, Steele J, et al. Twovariants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolardisorder. Molecular Psychiatry 2008;14:487e91.
Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, et al. Genome-wideassociation and meta-analysis of bipolar disorder in individuals of Europeanancestry. Proceedings of the National Academy of Sciences 2009;106:7501e6.
Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scalegenome-wide association analysis of bipolar disorder identifies a new suscep-tibility locus near ODZ4. Nature Genetics 2011;43:977e83.
Sklar P, Smoller JW, Fan J, Ferreira MAR, Perlis RH, Chambert K, et al. Whole-genomeassociation study of bipolar disorder. Molecular Psychiatry 2008;13:558.
Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, et al. CaV1.2calcium channel dysfunction causes a multisystem disorder includingarrhythmia and autism. Cell 2004;119:19.
Splawski I, Yoo DS, Stotz SC, Cherry A, ClaphamDE, KeatingMT. CACNA1Hmutations inautism spectrum disorders. Journal of Biological Chemistry 2006;281:22085e91.
Tabangin ME, Woo JG, Martin LJ. The effect of minor allele frequency on the like-lihood of obtaining false positives. BMC Proceedings 2009;3(Suppl. 7):S41.
Takata A, Kim SH, Ozaki N, Iwata N, Kunugi H, Inada T, et al. Association of ANK3with bipolar disorder confirmed in East Asia. American Journal of MedicalGenetics Part B: Neuropsychiatric Genetics 2011;156:312e5.
Tesli M, Koefoed P, Athanasiu L, Mattingsdal M, Gustafsson O, Agartz I, et al.Association analysis of ANK3 gene variants in nordic bipolar disorder andschizophrenia caseecontrol samples. American Journal of Medical Genetics PartB: Neuropsychiatric Genetics 2011;156:969e74.
Van Den Bogaert A, Sleegers K, De Zutter S, Heyrman L, Norrback K-F, Adolfsson R,et al. Association of brain-specific tryptophan hydroxylase, TPH2, with unipolarand bipolar disorder in a Northern Swedish, isolated population. Archives ofGeneral Psychiatry 2006;63:1103e10.
Weber H, Kittel-Schneider S, Gessner A, Domschke K, Neuner M, Jacob CP, et al.Cross-disorder analysis of bipolar risk genes: further evidence of DGKH as a riskgene for bipolar disorder, but also unipolar depression and adult ADHD. Neu-ropsychopharmacology 2011;36:2076e85.
Westfall PH, Young S. Resampling-based multiple testing. New York: Wiley; 1993.WHO. International statistical classification of diseases and related health problems
10th Revision. World Health Organisation; 2007.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of HardyeWeinberg
equilibrium. The American Journal of Human Genetics 2005;76:887e93.Williams HJ, Craddock N, Russo G, Hamshere ML, Moskvina V, Dwyer S, et al. Most
genome-wide significant susceptibility loci for schizophrenia and bipolardisorder reported to date cross-traditional diagnostic boundaries. HumanMolecular Genetics 2011;20:387e91.
Winokur G, Coryell W, Endicott J, Akiskal H. Further distinctions between manic-depressive Illness (bipolar disorder) and primary depressive disorder (uni-polar depression). American Journal of Psychiatry 1993;150:1176e81.
Winokur G, Coryell W, Keller M, Endicott J, Leon A. A family study of manic-depressive (bipolar-I) disease e is it a distinct illness separable from primaryunipolar depression. Archives of General Psychiatry 1995;52:367e73.
Wittchen HU, Höfler M, Gander F. Screening for mental disorders: performance ofthe Composite International Diagnostic e Screener (CID-S). Methods inPsychiatric Research; 1999:59e79.
Wray NR, Pergadia ML, Blackwood DHR, Penninx BWJH, Gordon SD, Nyholt DR,et al. Genome-wide association study of major depressive disorder: new results,meta-analysis, and lessons learned. Molecular Psychiatry 2012;17:36e48.
Xia Z, Storm DR. The role of calmodulin as a signal integrator for synaptic plasticity.Nature Reviews Neuroscience 2005;6:267.
Zhang X, Gainetdinov RR, Beaulieu J-M, Sotnikova TD, Burch LH, Williams RB, et al.Loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolarmajor depression. Neuron 2005;45:11.
Zhang Y, Meltzer HL. Increased content of a minor ankyrin in erythrocyte-membranes of bipolar subjects. Psychiatry Research 1989;27:267e75.